Last week, my Bento Lab co-founder Bethan and I spent four days in Norway for the 2019 International Barcode of Life conference (iBOL).

The annual conference focuses on DNA barcoding, a method of identifying species based on sequencing short snippets of DNA. We were invited to exhibit Bento Lab as an exciting option for accessible field research equipment. It was the first science conference we have attended in a while, and we were psyched to talk to everyone about the future of DNA barcoding, and how Bento Lab can contribute.
Check our Youtube channel for a short video blog to get a sense of the vibe of the conference, the pace of conversation at our exhibition stand, and the beauty of Trondheim in the summer.
What is DNA Barcoding?
DNA Barcoding is a method to identify species by analysing a specific region of DNA, the so-called ‘DNA barcode’ region, and comparing it to a reference library. The DNA region depends on the specific biological kingdom. For example, all animals are identified using the same specific DNA region, whilst all plants are identified using a different region.
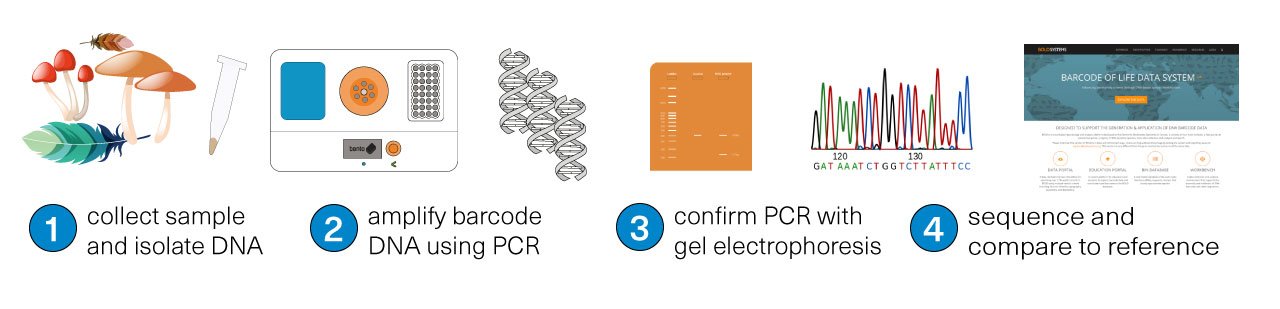
The first step is collecting the sample for study. Then, using a tool like Bento Lab, the DNA is isolated from the sample (1), and the target ‘DNA barcode’ is amplified using PCR (2), meaning that lots of copies of the physical DNA is made. The success of the PCR step can be confirmed using the method of gel electrophoresis (3). Finally, the DNA barcode is sequenced (4). The matching species can be identified by comparing the sequenced DNA barcode to reference databases.
The DNA can be sequenced by sending the amplified DNA to a sequencing service, or by using a portable DNA sequencing machine, like the Nanopore MinION (as some of Bento Lab’s users do) or the Illumina iSeq (like Genome Prairie, one of our communities in Canada).
Tools for field-ready sample preparation

During the four days, we had hundreds of intense conversations about the potential of using Bento Lab for mobile research. The iBOL conference participants are a range of scientists conducting research in a huge variety of settings, so there was a lot of excitement about the mobility and ease of travel with Bento Lab, and we came away with lots of ideas and excitement for the future. There was also a lot of discussion about supporting citizen science groups and school classes to make contributions to DNA barcoding and making a difference to biodiversity. This is also part of Bento Lab’s core mission.
Illuminate Biodiversity to change the way humanity understands our planet
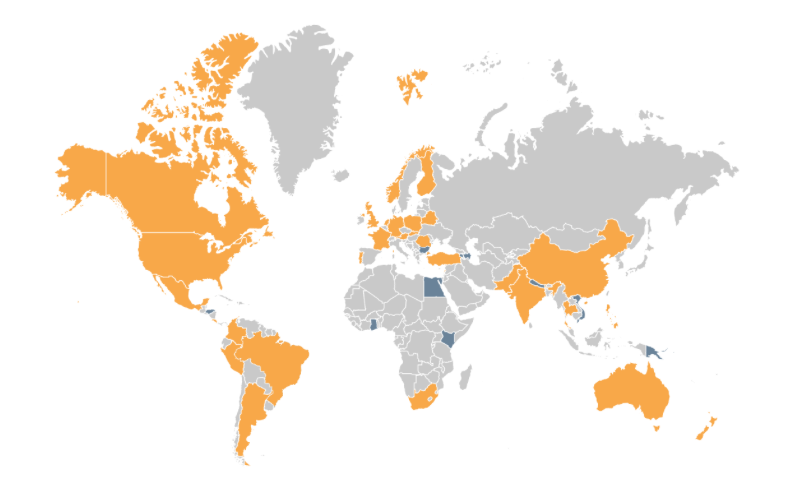
The conference organised by the iBOL consortium, a group of 30 member nations, plus 9 associate nations. All of the nations are supporting DNA barcoding activities in their region to increase the understanding of biodiversity, and shifts in species distribution. Their first major program, BARCODE 500K, finished in 2015 with the successful barcoding of the DNA of 500,000 species. Along the way, this helped demonstrate the value of DNA barcoding at scale, to build “a digital identification system for life”, as iBOL puts it.
The successor program, called BIOSCAN, launched this year. It focuses on three aspects:
- speeding up the discovery of species,
- investigating the interaction between species,
- and tracking shifts in species distribution.
When BIOSCAN is completed in 2025, it is planned to have extended to cover 2.5 million species.
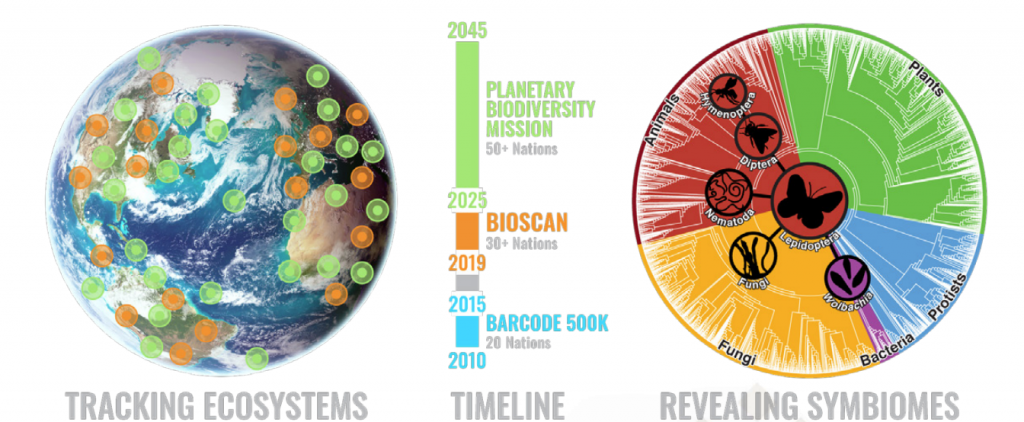
The ultimate scientific goal of iBOL is to build a planet wide biodiversity surveillance system, to deliver a complete and repeatedly updated map of multi-cellular life. This Planetary Biodiversity Mission, or PBM, is targeted to exist by 2045. Ultimately, iBOL is driven by concern for our planet, in the hope that by shining a light on the health of the biodiversity on the planet, we will be able to protect the planet to the benefit of all lifeforms.
Postscript

We had an amazing four days at the iBOL conference. Check out our video blog to get a sense of our time in Trondheim.
I was fortunate to be able to speak to some of the keynote speakers and organisers, including iBOL’s scientific director Paul Hebert, Dirk Steinke and Sujeevan Ratnasingham from the University of Guelph (the “birthplace of DNA barcoding”), as well as Ingrid Ertshus Mathisen and Stine Svalheim Markussen from Artsdatabanken (the Norwegian Biodiversity Information Centre).
Some clips from our conversations made it into the vlog, and we’ll post standalone interviews in the coming weeks. Subscribe to our YouTube channel, so you don’t miss them. I’m very grateful for the time everyone gave me – thank you very much.
Special thanks also to my friend Kazutoshi Tsuda from YCAM who was attending the conference. I have written about his amazing personal biotechnology workshop with Bento Lab and Nanopore MinION sequencing previously.




