What is DNA barcoding?
DNA barcoding is a method used to identify species. It works by analysing a specific region of DNA. This region is called the DNA barcode. The sequence of this DNA barcode is then compared to a reference library which contains information of many species linked to their barcodes.
There are different types of barcoding regions used for different biological kingdoms. For example, all animals are identified using the same specific DNA region, whilst all plants are identified using a different region.
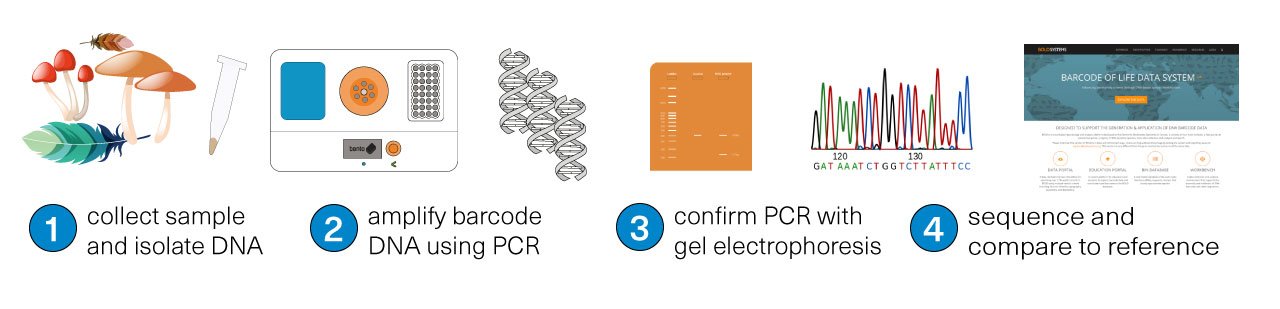
The first step is to collect a sample for study. Then, using a tool like Bento Lab, DNA is extracted from the sample (1) and the target DNA region is amplified using PCR (2), meaning that lots of copies of the DNA barcode are made. Visualisation of amplified DNA by gel electrophoresis (3) determines whether the PCR was successful. Finally, the DNA barcode is sequenced (4). The species can be identified by comparing the sequenced DNA barcode to reference databases.
The DNA can be sequenced by sending the amplified DNA to a sequencing service or by using a portable DNA sequencing machine.
We recommend you ensure that you have access to a sequencing solution (Sanger sequencing, or Oxford Nanopore MinION sequencing) before planning any DNA barcoding work.
You can find out more information, protocols, and guides in the drop down menu at the top of the page.
Are you interested in DNA barcoding?
We are passionate about making the power of PCR available to anyone. Using DNA barcoding to identify species can be essential for many reasons, from food safety to conservation.
If you are interested in carrying out this workflow for yourself, we would love to hear from you! Over the coming weeks and months we are adding to our Knowledge Hub of DNA barcoding resources. Let us know what would be most helpful to you!
What do I need to get started with DNA barcoding?
The reagents, consumables and equipment you would need to extract DNA from your sample and carry out a DNA barcoding protocol is as follows:
Reagents
Consumables
- Sample from fungus/invertebrate/bird/fish/mammal/plant
- Disposable scalpels or razor blades
- Nitrile gloves
- Paper towel
- 2-200μl Pipette Tips (1 rack)
- Microcentrifuge Tubes
- PCR tubes
- You will also need access to a Sanger sequencing service to sequence your amplicons
Equipment
- Bento Lab
- 250 mL beaker or conical flask
- 2-20μl Adjustable Pipette
- 20-200μl Adjustable Pipette
- Microwave
Scientific background: How does DNA barcoding work?
DNA barcoding relies on a region of DNA that varies significantly between different species to allow taxonomic differentiation, but that is flanked by regions of DNA that are the same between different species for PCR primers to bind to. A primer is a short sequence of DNA that is added to a PCR mastermix to indicate which region of the sample DNA is to be amplified. The forward primer binds to one strand of DNA, the reverse primer binds to the opposite strand in a different position and the section of DNA between the primers is amplified.
The DNA region used for barcoding differs between the kingdoms:
- In animals, the DNA barcode is within the gene for cytochrome c oxidase subunit 1 (CO1). Different sections of this gene are used to barcode phylum within the animal kingdom, such as invertebrates, birds, fish and mammals.
- In fungi, the most commonly used DNA barcode is the internal transcribed spacer (ITS) region. Either the entire ITS region is amplified or a subunit, depending on the fungal species of interest and the sequencing method being used.
- There are several candidates for DNA barcoding in plants. The two gene targets recommended by the Consortium for the Barcode of Life (CBOL) Plant Working Group are maturase K (matK) and ribulose bisphosphate carboxylase (rbcL).
DNA barcoding approaches can be used to identify a single species or a community of species.
Single species barcoding is when you collect a sample from a plant, fungus or animal, extract DNA from the sample, amplify the DNA barcode using PCR and send the DNA barcode for sequencing. An example of this is when mycology groups used Bento Lab to identify and rediscover UK fungal species as part of Kew’s Lost and Found Fungi project.
Community barcoding, or metabarcoding, is when a sample contains a mixture of species so DNA is extracted, amplified and sequenced from all the species in the mix that are targeted by the DNA barcode used. Examples of metabarcoding are profiling the fungal diversity in a soil sample (mycobiome), or finding out what types of plants honey bees forage on from the pollen in honey.
Why is DNA barcoding important?
There are many applications of DNA barcoding and reasons why it is important; for scientific discovery, for human health and for the health of our planet.
- Species discovery – the International Barcode of Life (iBOL) project seeks to make DNA barcoding globally accessible for the discovery and identification of all multicellular life on Earth.
- Detection of invasive species – DNA sampled from the environment (eDNA) can be barcoded to monitor the presence of invasive species of concern.
- Ecological assessment for conservation – understanding biodiversity and the effect of anthropogenic changes to make predictions for species survival and the planet’s future.
- Telling apart cryptic species – these are species that appear identical but are genetically distinct, which can have important implications for biodiversity, wildlife management and disease control.
- Diet analysis – it is increasingly understood that what we eat and drink affects our gut microbiota, and therefore our health. In a new era of personalised medicine, dietary advice could be tailored according to an individual’s gut microbiome (bacteria) and mycobiome (fungi).
- Food safety – ensuring that the food we are sold is properly labelled and contains only what it should. For example, DNA barcoding was used to discover horse meat in frozen burgers following suspicious movement of horses around Europe.
What will the results look like?
If you have successfully extracted DNA from your sample and amplified the DNA barcode with the relevant primers, you will see a single band on the gel for each sample. This means the PCR product(s) should be suitable to use for sequencing.
The amplicon can then be sequenced via a Sanger sequencing service or Oxford Nanopore MinION sequencer.
When the sequence has been produced, you can use the Basic Local Alignment Search Tool (BLAST) on the National Center for Biotechnology (NCBI) website to find the closest species identity match to your sample in their database.
