Cover picture: The Urra fieldstation in southern Spain. Credit: Katie Cook
Imagine yourself looking out across a desert landscape, with a warm breeze and the faint sound of busy insects in a dry landscape. This is the setting for the Urra Field Centre in Spain, where this April, Dr Elizabeth Duncan and her colleagues took a group of 40 students for a Mediterranean Ecology field trip.
Based at the University of Leeds, Dr Duncan studies the evolution of bees. There are two different groups: social bees and solitary bees. Social bees, such as honeybees and bumblebees, live in groups and depend on queen bees to reproduce. As their name suggests, solitary bees live on their own. They build burrows underground, or nests in buildings. Solitary bees make up 90% of the bee species in the UK, and bees, together with other pollinating animals, help to pollinate a third of all the food we eat. Like honeybees and bumblebees, solitary bees are under threat from changes to their habitat.

Included on the agenda for the Urra field trip: a project to identify Nosema infections of bees. A fungal parasite, Nosema invades the guts of the bee, and spreads through spores. When infected, bee colonies can fail to grow and dwindle away. For commercial beekeepers, this infection can mean a significant decline in colony productivity. The parasite also poses a risk to solitary bees, as the infection can spill over from commercial colonies into wild bee populations.
To an expert eye, the simplest method to identify a Nosema infection is under the light microscope. However, more accurate tests are possible using genetics. Researchers have developed methods based on PCR, which are sensitive enough to detect low levels of pathogen infection.
Loading the gel to look for evidence of Nosema in our bee samples using the @theBentoLab kit. @e_j_duncan #LeedsMedTrip pic.twitter.com/Dj0chqGLOu
— Chris Hassall (@katatrepsis) April 2, 2019
The sweet sound of science as our @theBentoLab kit whizzes into action, running a PCR on some bee tissue looking for evidence of parasites. Exciting incorporation of molecular biology into the #LeedsMedTrip by @e_j_duncan. Stay tuned for the results! pic.twitter.com/PpbbZfOB5O
— Chris Hassall (@katatrepsis) April 1, 2019
Throughout the week, the team collected honeybees from around Urra as samples for their barcoding experiments. To detect Nosema, the group needed to extract the Nosema fungal DNA, which is only found in the gut of bees. The students dissected the gut of the bees and extracted DNA from a very small portion. Using these samples, they ran conventional PCR assays and visualised the results, all with Bento Lab.
The PCR assay was designed to generate 2 products, the first to confirm the presence of honeybee DNA, and the second to search for the presence of Nosema DNA. In good news for the Urra region, the group found no presence of Nosema DNA in any of the samples they collected.
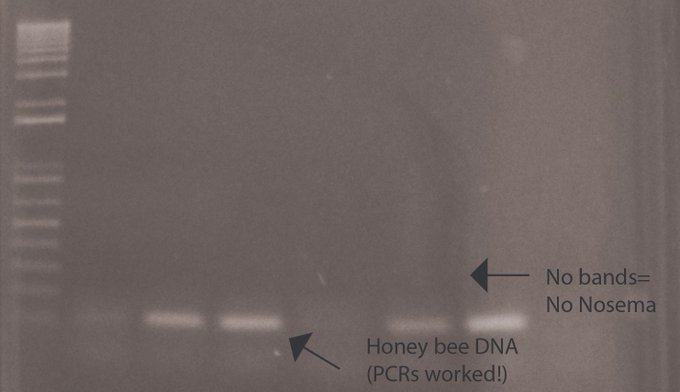
“And the results are in….. – one band for all our samples = Healthy honeybees! No signs of disease yet – good for Urra @katatrepsis @benitez_lab @BillKunin! Now to try more samples & experiments with our BentoLab #LeedsMedTrip” Credit: Elizabeth Duncan
When we spoke, Dr Duncan noted there is limited research available on solitary bees, as they are more challenging to work with. Given the commercial nature of social bees, there is more funding interest in honeybees and bumblebees. It’s also harder to collect wild bees for research, whereas samples of honeybees and bumblebees are easily available from breeders.
In the short-term, there are lots of simple ways that everyone can help wild bees, from growing a patch of wildflowers, to joining a recording scheme to keep track of local bees. If you are interested in supporting wild bees, take a look at Solitary Bee Week to learn more.
I never thought i would be able to do molecular biology in the field @UrraFieldCentre testing wild olive trees, rapeseed and wild rocket @e_j_duncan @katatrepsis , let you know the results later today pic.twitter.com/7UVPeyyjbK
— Benitez-Alfonso lab (@benitez_lab) April 3, 2019
Looking for bee pathogens round 2! #LeedsMedTrip pic.twitter.com/RsLPAeZzsU
— Elizabeth Duncan (@e_j_duncan) April 4, 2019
The field trip this April was a pilot run to test validate the field-based PCR assay. Along with a new group of students, Dr Duncan will be returning next year, and she plans to bring along tools for sequencing onsite for further research, including the portable sequencer MinION from Oxford Nanopore Technologies. Using these state-of-the-art molecular tools, Dr Duncan and future students will expand this work to detect other pathogens, and to measure biodiversity.




