Over the past few years, field mycologists (amateur fungal experts) in the UK have been using Bento Lab to help identify fungal biodiversity and characterise new species to the UK and to science.
Watch Bento Lab Co-Founder Bethan Wolfenden’s interview with Brian on YouTube.
Fungal biodiversity is one of the big frontiers in biodiversity exploration and conservation. Fungi belong to their own biological Kingdom distinct from plants and animals – a Kingdom that contains the more familiar mushrooms and moulds, but also an amazingly diverse set of other forms, such as brackets, puffballs, earthstars, crusts, spindles and coral fungi, cup fungi, earthtongues, leaf spots, woodwarts, earthstars, and a vast range of microfungi.

Kingdom Fungi is estimated to contain between 2.2 and 3.8 million species, over six times the estimated number of plants, but even after several centuries of study only 144,000 species have been formally described (given an officially recognised name). Currently around 2,000 new species are described per year. The gap between what we think is out there compared to what is known is enormous. In the report ‘State of the World’s Fungi’, the Royal Botanic Gardens Kew estimated that around 93% of fungal diversity on Earth remains to be discovered.
Fungi are an essential part of our natural world, as decomposers and recyclers, engaging in beneficial symbioses with most plants, and keeping plants, insects, and ecosystems diverse and in balance. They are also biodiversity in their own right, and just as important to protect and conserve as other organisms. However, without being to accurately identify fungi it is extremely difficult to either understand what they do, where they are, or how we can try to protect them from habitat loss, pollution, and climate change.
To help improve the identification of fungi, over the past few decades professional mycologists have developed a wide range of DNA-based methods to improve the identification of fungi. The most used method is DNA barcoding – the characterisation and identification of fungal species using sequences of species-specific regions of DNA.
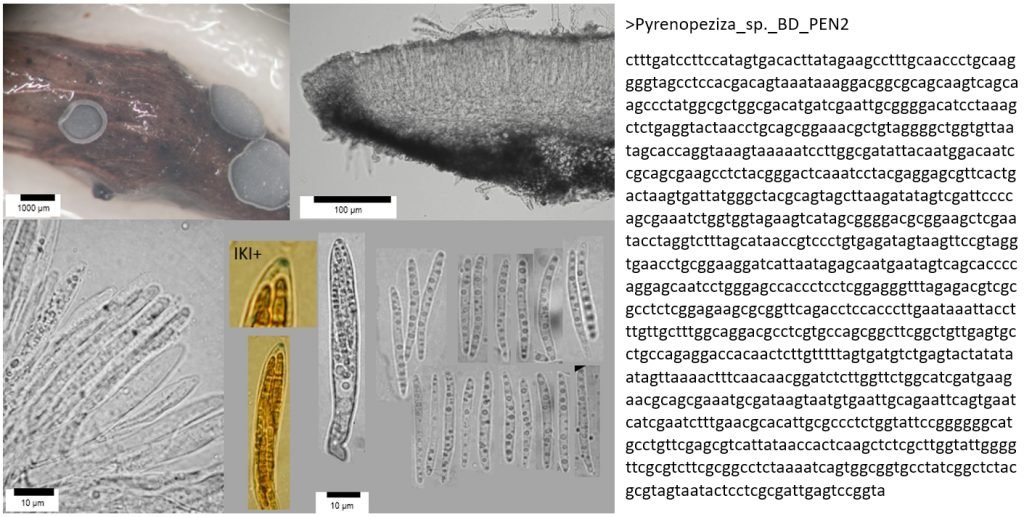
DNA barcoding has become a revolutionary methodology for the identification of fungi, following on from the invention of light microscopy in the 17th Century, scanning electron microscopy in the 1970s, and digital photography and social media in the 21st century. It is morphology-independent, which means it can work hand-in-hand with other identification methods to confirm or refute an identification; be used on its own to compare samples to reference material; and can also detect the same species from soils, plant tissues, water, or air samples. It relies on the assumption that different species have single distinct DNA barcode regions that can be differentiated from those of other species (which isn’t always true using standard barcode regions), but in general is an extremely useful tool for species discovery and identification.
However, a major obstacle to the effectiveness of DNA barcoding has been that it has been limited to a relatively small number of academic scientists across the world, and has been almost inaccessible to the much larger global community of fungus enthusiasts and field mycologists who lack both resources and knowledge to do this work. It has only been in recent years that this scientific divide has begun to be bridged, pioneered by a growing number of “DNA-enabled” members of the public, ranging from youth citizen scientists, to experienced field mycologists in the USA and other countries.
Bento Lab and UK Field Mycologists
In the UK, Bento Lab has been one of the main tools that has allowed UK field mycologists to join this movement.
Pembrokeshire Fungus Recording Network (PFRN)
In the UK, David Harries and the Pembrokeshire Fungus Recording Network (PFRN) were one of the first fungus recording groups to become involved in DNA barcoding of fungi. The group initially collaborated with Aberystwyth University Mycology Group in the collection of grassland fungi for barcoding of reference material for soil DNA work. After seeing the DNA barcoding process first-hand, David soon realised that it would be possible for the PFRN to start DNA barcoding specimens if he could source the equipment and reagents.

Members of the PFRN learning DNA barcoding at Aberystwyth University. © David Harries.

After initial work using donated or second-hand equipment, the group became one of the early testers of Bento Lab in 2017, and adapted a quick, safe, and easy “DNA dipstick” extraction protocol for use with fungi. The final step of the laboratory process was the DNA sequencing of PCR products, which was not possible to do at home, but could be arranged at Aberystwyth University. Sequence analysis skills were developed using a range of free software packages and online resources.
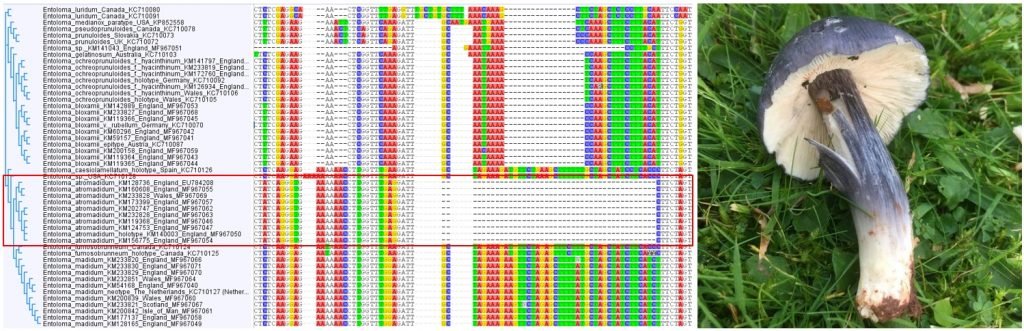
Once becoming confident in fungal DNA barcoding, the group began using this methodology to help unravel a number of known cryptic species complexes in Wales, including blackening waxcap mushrooms, and a DNA-based study of green and brown earthtongues in Wales, funded by Natural Resources Wales and the Lost and Found Fungi project (LAFF – see below for details of this project). Ongoing work is addressing some of the groups’ more interesting specimens, including poorly known or potentially undescribed species in habitats important for fungal conservation.
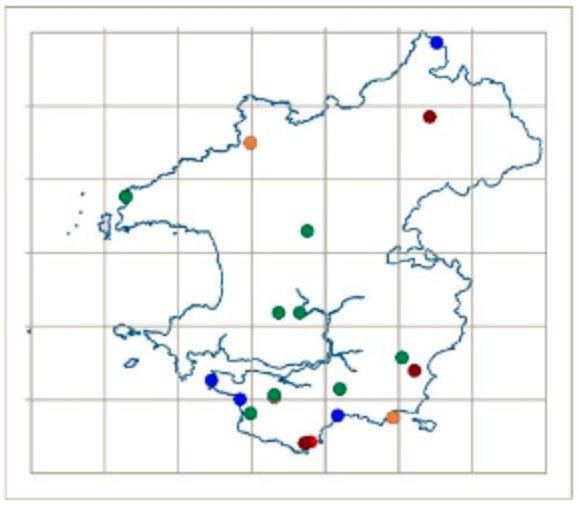


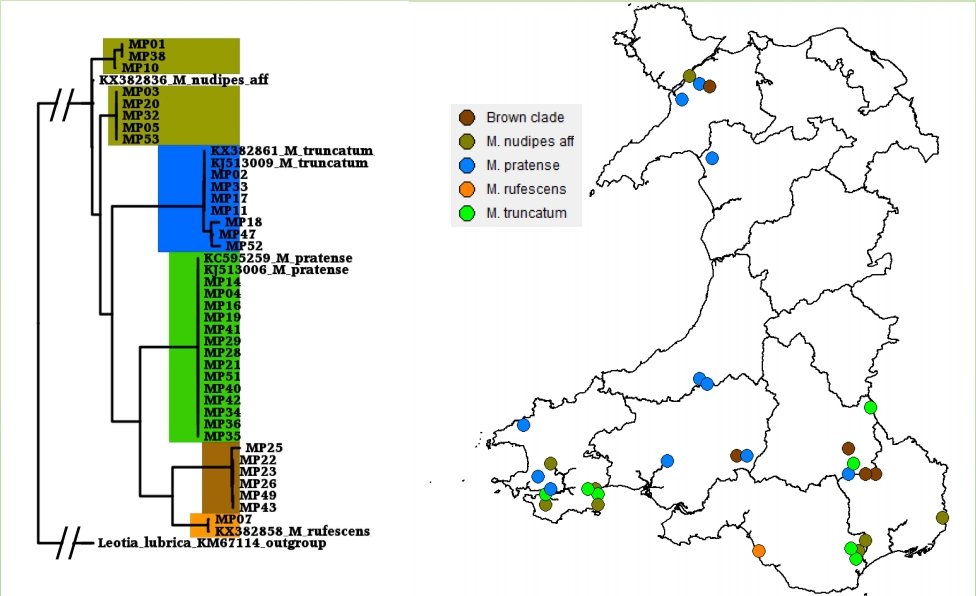
The Lost and Found Fungi project
The Lost and Found Fungi project (July 2014-March 2020) was a citizen science/volunteer orientated project co-ordinated by the Royal Botanic Gardens Kew, funded by the Esmée Fairbairn Foundation. Although the primary drive of the project was volunteer conservation surveying for a defined list of rarely recorded species, it also prioritised enabling and upskilling field mycologists to improve the UK’s capacity for conservation recording and identification. It soon became apparent that a growing number of individuals and groups within the UK fungus recording community, and especially many of the more experienced members, were extremely interested in DNA barcoding their most important finds.
To try to address this need, in late 2018 the LAFF project obtained three Bento Labs and began the process of teaching several fungus groups to use them for fungal ID, adding a further two Bento Labs in late 2019. The LAFF project ran workshops throughout 2019 to introduce DNA barcoding and DNA sequence analysis to interested groups throughout England, Wales, and Scotland; and conducted a joint fungus recording groups DNA workshop in partnership with the British Mycological Society.


A sequencing account was set up with the Core Genomic Facility in Sheffield University to clean and sequence participants’ PCR products using the project’s support budget, and the project provided support for sequence and taxonomic analysis for the generated sequences. Although the LAFF project was only able to provide this support for a limited time before the project ended, by the end of the project in March 2020 there were four fungus groups capable of independent DNA barcoding in the UK, and a further two following close behind. Two of these groups are profiled below.

Sussex Fungus Group
Nick Aplin of the Sussex Fungus Group was one of the first loanees of the LAFF project’s Bento Labs. Although Nick is an amateur expert in field mycology and microscopy he had no previous molecular biology background, but was keen to give the DNA barcoding process a try.

After a few introductory workshops and occasional guidance, plus a lot of independent work while the LAFF project was busy elsewhere, Nick soon picked up the basics and began successfully extracting and amplifying fungal DNA from a wide range of different types of fungi, ranging from large mushrooms to single microfungi fruitbodies less than 0.2 mm in diameter. Notable finds so far include confirming the identity of species of conservation concern; identifying new species of fungi to Sussex, to Europe, and potentially to science; and also matching up asexual and sexual stages of fungi.
Hampshire Fungus Recording Group
The Hampshire Fungus Recording Group is a long-established group populated by very expert and experienced mycologists, regularly making important finds from diverse habitats within the New Forest and surrounding areas. Between them the members possess a wide range of laboratory and technical and computer skills, although none had previously undertaken DNA barcoding work. Starting off with a series of LAFF-run workshops in November 2019, under the lead of Eric Janke the group is now independently producing PCR amplicons and sequences, and has recently confirmed several new species to the UK. Focusing on some of the more difficult fungal groups, they have also been finding previously unsequenced and poorly named species that require further taxonomic work for confident identification.

Amateur fungal DNA barcoding in the UK is still at a very early stage, but successes so far suggest that this work will continue to develop and expand. Several groups are now actively seeking funding to support further sequencing work. The processes of molecular identification can be complicated, but are easily achievable for the amateur mycologist willing to invest enough time into learning molecular techniques.
Hopefully this will help usher in a new wave of UK fungal DNA barcoding and biodiversity discovery, led by amateur experts and fungus enthusiasts rather than just by professionals. After all, it’s much easier to teach experienced field mycologists DNA barcoding methods than teach a molecular biology student how to morphologically identify fungi!
If you’re interested in becoming a field mycologist in the UK, the British Mycological Society has a list of regional groups on their website, and also a very active Facebook Group.



