For those interested in the potential of PCR to combat foodborne illnesses and zoonotic diseases, Marin et al. (2022) present an innovative study where a portable sequencing kit — combining Bento Lab and the MinION nanopore sequencer (Oxford Nanopore Technologies) — was used for on-site detection and characterization of Campylobacter and other bacteria in approximately five hours.
“This method could provide a basis for future studies to help farmers implement active zoonotic bacterial screening in real-time diagnostics to take measures that will reduce the prevalence, which is indispensable for active surveillance.” – Marin et al. (2022)
The Challenge: Food Safety & Rapid Detection
Campylobacter is one of the most important foodborne pathogens globally, particularly in poultry. Early detection of Campylobacter outbreaks in broiler flocks could significantly reduce the risks of human infection. However, due to the rapid production cycle of broiler chickens (from egg to supermarket in ~43 days) and the scale of poultry farming, outbreaks need to be identified using fast, practical, and cost-effective methods.
Unfortunately, current methods of Campylobacter enumeration using microbiological culturing are time-consuming and can take up to five days of work. This delay from sampling to results is not ideal given the rapid production cycle and the fact that broiler chickens can be on supermarket shelves within 24 hrs of slaughter. Faster, sensitive, and cost-effective solutions are urgently needed.
The Study: A Proof-of-Concept Workflow
In this study, the authors (Marin et al., 2022) proposed and tested a proof-of-concept same-day, on-site Oxford Nanopore sequencing workflow to detect Campylobacter in broiler chickens, using Bento Lab as a portable PCR workstation and ONT MinION for sequencing.
The study involved 42 broiler chicks, with 20% being orally infected with Campylobacter. Caecal (gut) samples were taken at two key points:
- Day 14 – When the broiler immune system matures
- Day 42 – When the birds are taken for slaughter
Two detection workflows were compared:
- Standard microbiological culturing (ISO/TS 10272-2:2017)
- Portable Bento Lab & MinION sequencing workflow
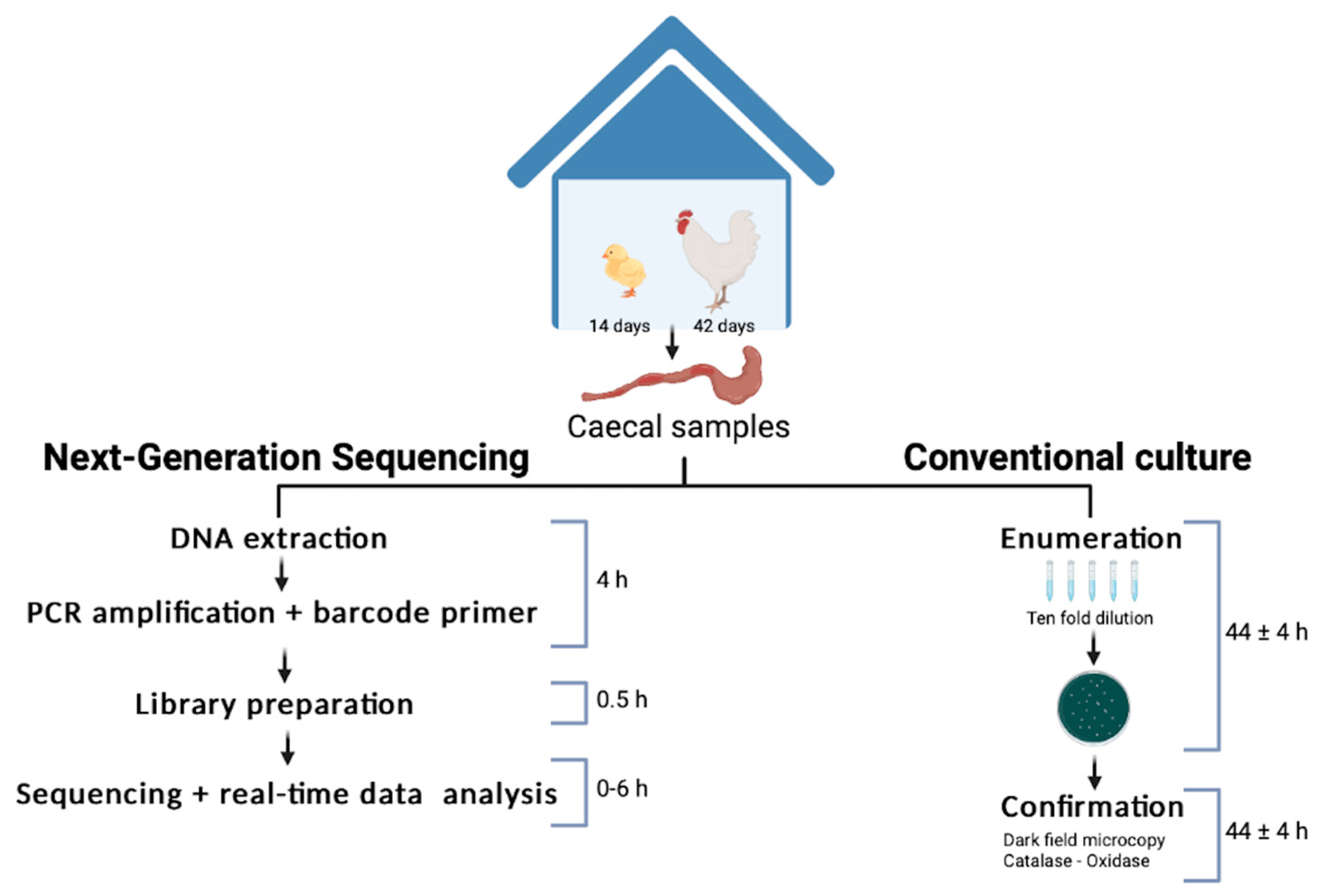
Timeline for the rapid method developed at farm level compared with the current culture detection method. Image CC-BY-4.0 the authors of Marin et al. (2022).
Bento Lab was used for
- DNA extraction from caecal contents using the Wizard Genomic DNA Purification Kit, Promega)
- Visualisation of DNA concentration and quality via agarose gel electrophoresis
- Library preparation for MinION sequencing
Following library preparation, the libraries were run on a MinION flow cell for six hours, with reads logged every 10 minutes to allow determination of the shortest amount of time needed to generate sufficient data for analysis. Raw reads were base-called using Albacore 2.1.7, poretools 0.06, and reads were assigned to taxonomic identities using RDP classifier.
The results showed that:
- Campylobacter was detected in all caecal samples, but it was absent in environmental samples and day-old chicks, demonstrating the sensitivity of the workflow
- The culturing ISO/TS 10272-2:2017 method took four days from start to finish, whereas the Bento Lab/MinION process produced actionable data within five hours.
- The Bento Lab/MinION workflow allowed simultaneous detection of Campylobacter as well as an assessment of bacterial taxonomic diversity and relative abundance present within the caecal samples, showing clear differences between the two sampling times.
The authors calculated that costs per sample for MinION sequencing were ~18x more expensive than the conventional culturing method. However, this could be reduced per sample by increasing the throughput, since ONT sequencing becomes significantly cheaper as sample numbers increase. The authors also didn’t consider additional costs of staff time.
Marin et al. (2022) concluded that “[we] have demonstrated that MinION-based workflow at the farm level is viable and highly accurate by comparing it with ISO/TS 10272-2:2017.”
Overall, this looks like an amazing application of portable PCR diagnostics for zoonotic disease detection, and one that could be applied in many more areas of the food production industry!
You can read the article here:
Marin et al. (2022). Rapid oxford nanopore technologies MinION sequencing workflow for Campylobacter jejuni identification in broilers on site—a proof-of-concept study. Animals, 12(16), 2065.


