Hey PCR enthusiasts!
Here are this week’s highlights from Bento Lab.
Bento Lab in the Field
The curious case of the jelly fungus with snake-like spores

We’re very happy to share this great article by field mycologist Nick Aplin (of Sussex Fungus Group, United Kingdom), on the discovery, characterization, and taxonomy of a tiny obscure jelly fungus, Stypella mirabilis (now Myxarium mirabile), DNA barcoded using Bento Lab 🙌.
Tiny jelly fungi are often more poorly investigated than the larger fungi. In this particular case, Nick was surprised and intrigued by the unusual snake-like spores of his collection. After DNA barcoding it using Bento Lab and the filter-paper DNA extraction method, the sequence didn’t match anything well in DNA sequence databases. But he suspected it could be a described species that hadn’t yet been sequenced.
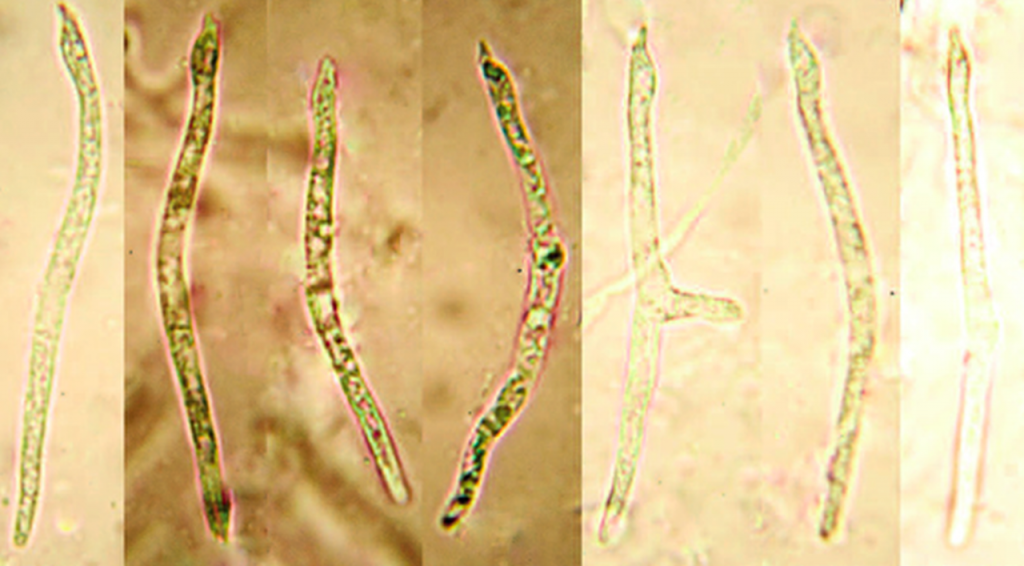
After correspondence with Dr. Peter Roberts, a world expert on the jelly fungi, it turned out that this collection was identical to a species he had described back in 1998, Stypella mirabilis. The epithet “mirabilis” comes from the Latin “to marvel at” and “wonderful”, reflecting the fact that he was also surprised and delighted by the spores. It was also in his opinion a very rare species, a suspicion supported by the lack of UK or global records between its description and Nick’s rediscovery. The DNA barcode indicated that it now would be better placed taxonomically in the genus Myxarium (along with many other species previously placed in Stypella), so Nick formally proposed the name of “Myxarium mirabile”,
Nick’s article expanded the knowledge of this extremely rarely seen fungus; provided a first DNA barcode; and improved its taxonomic classification.
Amazing work for someone doing the work for fun and personal scientific interest, with a microscope and Bento Lab in a spare room!
You can read the open access article here.
Interested in DNA methods and workflows? Subscribe for monthly insights.
Accessible PCR Spotlight
Identifying animals using DNA from their scat
We really love the idea of portable PCR applications becoming useful tools for ecologists and citizen scientists in surveying, monitoring, and identifying biodiversity. DNA barcoding is one application which many people find valuable for discovering and recording biodiversity, such as fungal barcoding. But what about other applications, such as specific PCR assays for particular animals?
We took a look at the feasibility of using Bento Lab to identify bats from their droppings, and thanks to some amazing publications by researchers at North Carolina University and elsewhere we think this would be very achievable, either with standard barcoding or with Oxford Nanopore MinION metagenomics. You can read our thoughts on this, with links to their articles, here.
Specific DNA assays for different target species would also be very possible, and we chanced upon a very nice article by Wilkinson et al. (2022) that used a workflow that allowed identification of arctic and red foxes from their scat using agarose gel electrophoresis. Their workflow could also broadly identify what they had been eating! You can read this article here. We would love to explore this kind of application further if anyone is interested in doing it with Bento Lab!
Paper Highlight
A new strategy for sexing parrots by Kroczak et al. (2021)
Parrots are an important group for veterinarians, bird breeders and conservationists, and knowing their sex is important for a range of bird health, breeding, and ecological research reasons. But they are often difficult to sex without using DNA, especially when very young. Unfortunately, parrots are quite genetically diverse. There is no single primer set that allows reliable sexing for all species, and information on which primers work best with specific parrot species is sparse and scattered in the literature.
We recently wrote a short note on a great article by Kroczak et al. (2021) who developed a new strategy for sexing the order Psittaciformes, using multiple primer sets and a strategy of using the most generally successful primers first to confirm results. The authors reported the results of these primers on male and female birds of 135 species of Psittaciformes, so users of the strategy will know exactly what to expect in terms of banding patterns and amplicon sizes.
You can find our note on the article here.
We don’t yet stock these primers (…yet? Let us know if you would like them). But we would be interested in hearing if there was any demand for these from any PCR enthusiasts or bird breeders!


