Hey PCR enthusiasts,
Here are some updates from Bento Lab!
Methods and Techniques
Fast 2-Step PCR (VPCR)
Do you need faster PCR for detecting DNA targets using standard PCR equipment and polymerases? If you do, you may be interested in a two-temperature PCR method that could reduce PCR run times to as little as 30 minutes or even 8 minutes!
The method by Chen et al. (2019) is called “VPCR” (not to be confused with V-PCR or “viability PCR”), based on the “V-shape” of the thermal cycling program. It involves thermal cycling rapidly between two temperatures rather than three, with zero hold time durations.
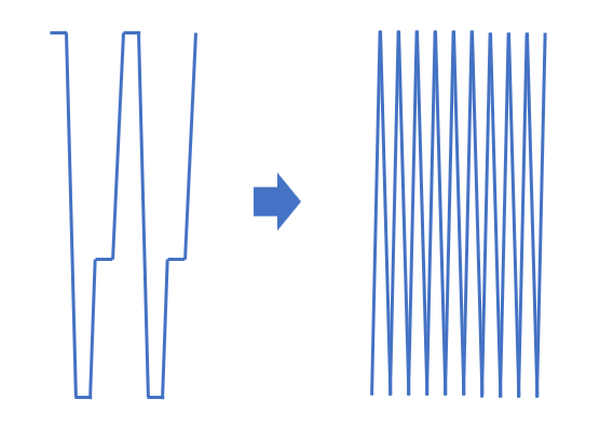
The denaturing, annealing, and extension steps occur during the dynamic heating and cooling processes instead of during temperature hold steps. Further time savings can be produced by also lowering the denaturing temperature and raising the annealing/extension temperature as amplicon concentration increases during the cycling.
Chen et al. (2019) found that VPCR could amplify a 500 bp amplicon in 16 minutes using a “fast” polymerase (KAPPA2G Robust), and in 32 minutes using a standard Taq polymerase. Shorter fragments amplified even more easily, and the authors managed to decrease the required time to amplify a 98 bp fragment to only 8 minutes!
The authors claimed no compromise in amplification efficiency or specificity for shorter fragments, which also means the method can be used for quantitative PCR (qPCR).
There are some disclaimers, in that success will probably vary depending on the specifics of the PCR application; the method may be best suited to shorter amplicons; and it may be easier to optimise for routine genotyping with standardised DNA extractions. We also haven’t yet tested this method ourselves.
Also, for use with Bento Lab, you will be limited by the shortest programmable temperature step being 5 seconds, which sets a minimum PCR time of around 25 minutes. However, this may be advantageous to some applications, and it’s still an amazing time saving if it works for your application.
You can find the article here:
Further examples of this method can be found in a later study of Pinelliae rhizoma (a chinese herb) by the same authors, and in a rapid genotyping study of tomato by a different group (subscription access only).
If you try VPCR, or are already doing something similar to it, and would like to share your experiences, please let us know!.
Product Highlight
Dipstick DNA Extraction Kit
If you want to rapidly extract and clean DNA for PCR from a wide range of tissues, then you may be interested in the ingenious filter paper dipstick method developed by Zou et al. (2020). It’s a great option for beginners, field-work, and low-throughput DNA extraction at an affordable price and using safe reagents.
You can rapidly extract PCR-quality DNA from a wide range of tissues in only 3 steps. The method involves grinding tissue in a lysis buffer, binding of DNA onto a filter paper dipstick, a wash step, and direct transfer of DNA into a PCR mix, all in 30 seconds!
We love this method because it’s very effective for some difficult tissue types, including fungi, lichens, and plants; and because it’s an easy method for anyone to do whether they’re beginners or more experienced.
Manufacturing dipsticks and solutions yourself can be difficult and time-consuming, so we’re very happy to be able to offer our Dipstick DNA Extraction Kit, which includes enough dipsticks and buffers for up to 100 extractions for $38.50. You can find it in our store here.

Methods and Techniques
Nucleic Acid Preservation Buffer (NAP Buffer or homemade “RNALater”)
Did you know that you can make your own DNA and RNA preservation solutions at a considerably lower cost than commercial solutions? Here is one recipe for a cheap but effective DNA and RNA preservative solution for animal tissues, their associated microbiomes, and possibly other tissue and DNA sources as well!
The solution has been called “Nucleic Acid Preservation” (NAP) buffer, and it appears to be based on the same chemistry as the popular RNALater™ preservation solution. To make 1 L all you need 7.44 g of EDTA, 7.35 g of sodium citrate tri-sodium salt dihydrate, and 700 g of ammonium sulphate in 1 L of water, adjusted to pH 5.2 using sulphuric acid. The method is more fully described in the article below:
The authors reported that samples stored in NAP at ambient temperatures contained high-quality DNA for up to 10 months, and tissue DNA concentrations were significantly higher in NAP than in tissues stored in 95% ethanol or cryopreserved. RNA losses were higher but there was no significant difference between the performance of NAP and a commercial RNA preservation solution (RNALater™).
The solution appears to be relatively rarely cited compared to commercial alternatives (e.g. RNALater), but it has been used for quite a few diverse applications, including sheep dung microbiome studies, chicken gut microbiome metagenomics, equine faecal microbiota, preserving deer organs for virus detection, and tree bark biofilm metabarcoding,
The chemistry is very similar or identical to that of RNALater, and you can find the now expired RNALater patent here: https://patents.google.com/patent/EP1657313A2/en.
The chemicals needed to make NAP buffer are inexpensive, safe, and widely available, except for the sulphuric acid for pH adjustment. And it seems quite simple to make if you have the chemicals, a pH meter, and some sulphuric acid to adjust the pH. So, if you need a cheap DNA and RNA preservative solution, then maybe this is one to try out!


