Hey PCR enthusiasts,
Here are some updates from Bento Lab!
Methods and Techniques
Line-PCR For High-Throughput Plant Genotyping
Line-PCR, developed by Lynagh et al (2023), is a rapid and cost-effective method of direct PCR for plant leaves that uses a minimal amount of plastic per sample. It involves poking a hole in a fresh leaf with a 1–5 mm piece of fishing line and putting the line fragment directly into a PCR. While poking the leaf, just enough DNA is captured as a smear on the plastic surface to use for PCR, but not so much leaf tissue that it contains too many PCR inhibitors.
Line-PCR has been shown to work well with normal PCR master mix (with no modifications), for Arabidopsis, rice, potato, and false flax (Camelina sativa). It’s sufficiently quick and easy that large numbers of plants can be sampled in the greenhouse or the field in one session, making it ideal for high-throughput genotyping. The line can also be stored in sterile distilled water for up to 24 hrs prior to PCR with no observed lack of sensitivity, allowing PCR master mix and primers to be added when back in the lab.
In the same study, the authors also developed a similar method called “CutTip” where pipette tips are used instead of fishing line, and the end of the tip is cut off into a PCR tube. This method is not quite as easy or efficient but could be done if no fishing line is available at time of sampling.
As with all direct PCR methods no DNA extract is retained for repeats or different loci, and this should be taken into account if a DNA extract is needed.
The article, and videos of Line-PCR in action (in the supplementary data), can be found here:
Product Highlight
Bento Lab Carry Case
Taking your Bento Lab on the road? Get peace of mind on the move with our durable carrying case!
Built tough to military-grade specifications, the case shields against water and dust, ensuring your lab stays safe even if it is briefly immersed in water.
We stock two sizes: one snugly fits your Bento Lab, while the larger option can fit extra equipment like pipettes so you can keep everything organized for easy travel.
You can find the Bento Lab carry case in our online store here.

If you have any other great products you’d like us to stock, please drop us a message!
Methods and Techniques
Rapid MinION Workflow for Antimicrobial Resistance Gene Detection using Bento Lab
Bloemen et al. (2023) recently developed and optimised a three-hour workflow that allows DNA extraction and Oxford Nanopore library preparation for bacterial metagenomics for portable on-site use. The authors developed the workflow to detect antimicrobial-resistant genes present in chicken fecal samples as a test case, but they believe it could also be used for other complex sample types.
Their final portable on-site method (illustrated below by the authors) involved:
- Cell lysis using the battery-powered Omnilyser X beat-beater
- Zymo Quick-DNA HMW magbead DNA purification
- AMPure XP bead purification with 0.4 bead/sample ratio
- Bento Lab Pro for all heating and centrifugation steps
- Nanopore sequencing library preparation using Oxford Nanopore VolTRAX
- Oxford Nanopore sequencing
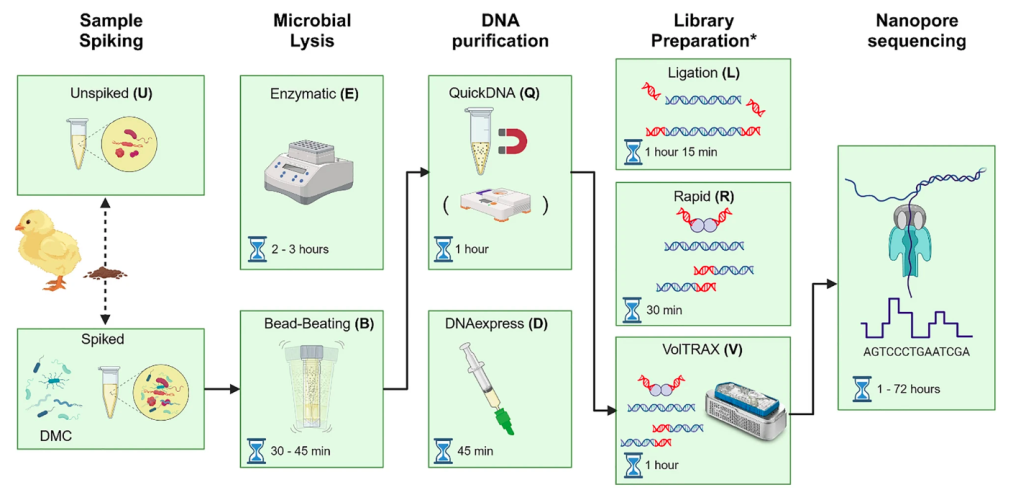
The method returned comparable amounts of high-purity DNA compared to their laboratory-based workflow involving enzymatic lysis and Quick-DNA HMW magbead extraction, albeit with shorter fragment sizes (27 kbp vs 58 kbp).
Importantly, the authors also used a defined mock bacterial community to spike some of their samples. This allowed them to determine the effectiveness of different extraction steps (for example enzymatic lysis vs beat-beating), and to identify the strengths and weaknesses of the different DNA extraction treatments in terms of bacterial groups detected.
An interesting tweak was to reduce the Omnilyser X bead-beater battery voltage from 6 V to 1.5 V, providing a much gentler beat-beating action that doubled the fragment length of extracted DNA. While this did reduce DNA yield by nearly 50% it significantly improved the sequencing results.
You can find the article here:


