Hey PCR enthusiasts,
Here are some updates from Bento Lab!
Software and Applications
GelAnalyzer 23.1: Free Electrophoresis Gel Analysis Software
Electrophoresis gel analysis software can be useful to to improve estimates of DNA fragment size and concentration in electrophoresis gels. But what software can you use to do this?
One great option is GelAnalyzer 23.1, a freeware software application written and released by Dr. Istvan Lazar Jr. and Dr. Istvan Lazar Sr.
GelAnalyzer was first released in 2010 and since that time has been used in over 1K scientific publications according to Google Scholar.
Its core features include:
- Analysis of gel images from many image sources
- Rotation and cropping of images
- Auto-detection of lanes
- Auto-detection amplicon peaks
- Full manual control of adding/modifying lanes and bands
- Background and distortion correction
- Quantitative and qualitative calibration and evaluation
It’s free, works on Windows/Mac/Linux, it’s relatively easy to use, and it has a good walkthrough guide on the website, so why not give it a go?
You can find it at http://gelanalyzer.com, and the official reference is: GelAnalyzer 23.1 (available at http://gelanalyzer.com) by Istvan Lazar Jr., PhD and Istvan Lazar Sr., PhD, CSc.
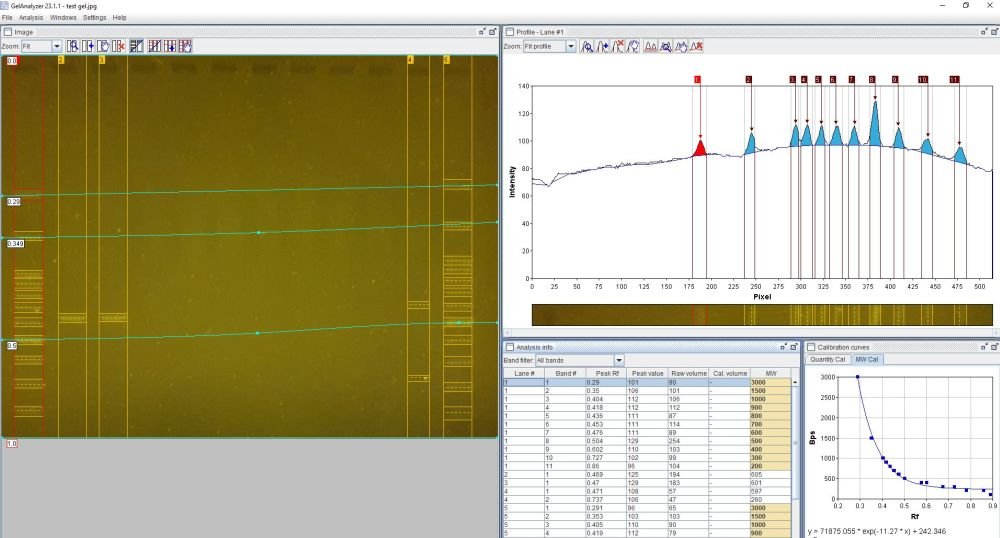
Below is an example of GelAnalyzer’s interface in use, showing detection of calibration lanes (the DNA ladder), and estimation of the length of sample amplicons in comparison to the DNA ladder:
Bento Lab Feature
Lab@Home vlog
Are you curious about setting up a PCR lab to work from home, and wondering what it would involve?
Take a look at our Lab@Home vlog series, where Dr. Jenny Shelton demonstrates DNA analysis on a kitchen table.
In the series you find out what molecular biology at home REALLY looks like, and learn about core “PCR-at-home” skills, including how to consider sterility for a temporary lab on a kitchen table.
If you have ideas for experiments you want to see in the future, please let us know! We love making resources that enable anyone to do the PCR work they want to do, wherever they want to do it.
You can find all of our Lab@Home vlog series on the Bento Lab website here.

Methods and Techniques
Super-High Throughput DNA barcoding with ONT MinION
What’s the highest throughput and lowest cost for DNA barcoding that you can imagine being possible today?
In a new preprint, an amazing 100K invertebrate samples were sequenced in a single run using Oxford Nanopore MinION sequencing! The authors also demonstrated the sequencing of 10K and 2K samples using smaller and cheaper Flongle flow cells.
The authors, Hebert et al. (2023) from the Centre for Biodiversity Genomics, University of Guelph, Canada, examined COI DNA barcode recovery from 100K malaise-trapped arthropods, testing cost and effectiveness in batches of 2K, 10K, and 100K samples.
To keep costs down while sample numbers increased, the researchers used:
- The latest MinION and Flongle flow cells (with R10.4.1 and Q20+ chemistry)
- Unique Molecular Identifier Tags to allow massive-scale multiplexing of PCR amplicons
- Small volume PCRs (12 µL and 6 µL)
- Cost-effective but high-throughput DNA extraction methods
The authors compared their results to PacBio Sequel sequencing of the same extraction set, allowing the direct comparison of barcode production efficiency and sequence fidelity between the two different technologies.
Their barcoding workflow results across the three sample scales showed:
- 87%-92% barcoding success
- 98% and 99% samples with less than 2% divergence between MinION and Sequel PacBio barcodes
- 94%-96% of sequences identical to Sequel PacBio barcodes
The authors calculate the sequencing costs at only $0.05/sequence at the 2K scale, and $0.01/sequence at the 100K scale. Importantly, this approach requires significantly smaller capital costs than other technologies, making it very well-suited for wide application around the world!
You can find the article preprint here:
Hebert et al. (2023). Barcode 100K Specimens: In a Single Nanopore Run. bioRxiv, 2023-11.


