Watch our interview with Ineke Knot on our YouTube channel.
Ineke Knot is bringing local health monitoring of great apes into the genomic age. Over the course of the last four years, she has developed methods to identify different types of nematodes using a fully portable system. Her research has recently been published in an open access paper: DNA Barcoding of Nematodes Using the MinION.
Hooray! It’s here: my first first-author publication! DNA barcoding of nematodes using the @nanopore #MinION and @theBentoLab. It’s #OpenAccess, follow the link to read the full article. If you’re interested in a summary, keep reading this thread. https://t.co/ww9tgXbgzF 1/12
— Ineke Knot (@ieknot) May 9, 2020
Protecting Great Apes from Infections
As our close genetic relatives, primates are susceptible to human infections. When primate communities are in contact with humans – either local people, researchers or ecotourists – they are vulnerable to the transmission of infections. Infectious diseases can decimate local populations of great apes. In 2006, an outbreak of Ebola led to a gorilla population decline of about 83%.
“Humans and great apes are so closely genetically related, that a lot of our diseases that we carry can also be transmitted to great apes.”
Ineke Knot
Like humans, apes are social animals. But as Ineke points out, how can you ask a gorilla to practice social distancing? This makes real time health monitoring incredibly important, so that other measures can be implemented to protect the primates.

Using Portable Genomic Tools to Identify Nematodes
As part of a PhD with the University of Amsterdam, Ineke has been developing DNA-based field methods to identify parasites in great apes. Her research focuses on nematodes – microscopic worms that include several parasitic species.
Traditionally, most zoonotic studies focus on viruses. Viral RNA evolves rapidly, and allows researchers to compare changes between a single generation. But working with viruses is notoriously difficult. RNA is more unstable and susceptible to degradation than DNA, which poses extra challenges when working in the field.
To demonstrate proof of concept, Ineke has focused her attention on nematodes to apply portable genomics to conservation.
When I started my PhD, I realized that nematodes are a kind of neglected tropical disease. In a lot of areas of the world, parasite infections are still really widespread health problems.
Ineke Knot
Ineke ran tests with four different species, Anisakis simplex, Panagrellus redivivus, Turbatrix aceti, and Caenorhabditis elegans.
To identify the species with DNA barcoding, the team chose a common barcode for nematode species identification: a gene fragment from the 18S ribosomal RNA gene that is about ∼900 base pairs in length. This test could be applied to samples from a range of different environments, from faeces, to soil, to marine sediments.

Sumatran Orangutan Conservation Programme (SOCP)
Last year, Ineke travelled to Indonesia to share her knowledge with the Sumatran Orangutan Conservation Programme. The NGO focuses on reintroducing orangutans to Bukit Tigapuluh National Park, an Indonesian forest where the species had been missing for over 150 years. As part of their rescue and rehabilitation work, the programme hopes to start using portable genomic tools like Bento Lab to study the microbiome of orangutans in their care.
Right now, the SOCP is fundraising in order to protect the orangutans from Covid19. If you are interested in helping, you can find more information on their fundraising website.


Training Local Staff and Spreading Knowledge
Providing training for the team in Sumatra has changed Ineke’s perspective. She is now on a mission to share her knowledge, and support wildlife conservationists that want to undertake molecular biology on remote field sites, like the Sumatran Orangutan Conservation Programme.
If you are interested to learn more about using portable genomics in the field, Ineke will be running a free tutorial on 13th August with WILDLABS, a community platform for conservationists and technology experts.
You can keep up with Ineke by following her personal blog, and her Instagram. Ineke also talks about science on Twitter as @ieknot.
“The underground world is an unexplored frontier, yet to be discovered.”
Dr Josef Vuch
Dark, dank and deep. While the mysterious depths of caves seem uninhabitable, they are home to a myriad of creatures, including bats, salamanders, insects and crustaceans. One such cave dweller is the Olm (Proteus anguinus) which also goes by the nicknames of “baby dragon” or “human fish”. A groundwater salamander found across Eastern Europe, the Olm recently join a display in public for the first time in Slovenia, as part of an exhibit of the many creatures living in the dark.

During the week, Dr Josef Vuch works as a molecular biologist in a biomedical research laboratory at the Children’s Hospital of Trieste. In his free time, he explores biospeleology research with the speleological section of the Club Alpinistico Triestino (Triestine Mountaineering Club). Biospeleology is the biology of caves – biospeleologists study the ecosystems of caves and the inhabiting organisms.

Caves are in constant darkness with high humidity. Nutrients are scarce, and many caves contain mixtures of potentially lethal gases. Yet, in this hostile environment life has adapted to survive without light and scant food, explains Josef. Crustaceans and insects have adapted; losing their sight, developing a cuticle to prevent dehydration and elongated appendages to detect space in the absence of light.
The unique environment of caves makes the life surviving there challenging to research. Josef notes, “The hypogean environment is not well studied because of its inaccessibility, and lack of commercial interest.” This means a vast number of species have yet to be identified, and turns the underground world into an unexplored frontier, yet to be discovered.

In the last decade, researchers have started implementing molecular techniques, such as DNA barcoding, to identify and characterise novel species. For example, researchers demonstrated the use of molecular data for taxonomic identification of Zospeum, and identified several new species. One advantage that DNA barcoding presents is the ability to use non-invasive methods to identify species by isolating DNA.

DNA barcoding relies on the standardised use of a short section of DNA from a specific gene, known as a “barcode region”. These barcode regions are chosen from highly conserved regions of the genome. This means less variation of the sequence within a species, allowing researchers to use the fragment for species identification. The most commonly used barcode region for animals is a segment of the cytochrome c oxidase I (COI) gene, which can be found in mitochondrial DNA. Dr Josef Vuch plans on using Bento Lab to extract DNA from samples and run PCR amplification of the barcode region, to produce amplicons for Sanger sequencing and analysis. Josef commented, “as a very compact instrument, Bento Lab is ideal for domestic or field use.”
“By amplifying the COI gene, it will be possible to molecularly identify animal species living in caves even from their remains or from environmental samples and discriminate individuals of the same species from different species, by measuring the genetic distance.”
Dr Josef Vuch

For those located near Trieste, you can find the Club Alpinistico Triestino online. Otherwise find a group near you by having a look for your national speleological organisation, which are listed by the International Union of Speleology.
Over the past few years, field mycologists (amateur fungal experts) in the UK have been using Bento Lab to help identify fungal biodiversity and characterise new species to the UK and to science.
Watch Bento Lab Co-Founder Bethan Wolfenden’s interview with Brian on YouTube.
Fungal biodiversity is one of the big frontiers in biodiversity exploration and conservation. Fungi belong to their own biological Kingdom distinct from plants and animals – a Kingdom that contains the more familiar mushrooms and moulds, but also an amazingly diverse set of other forms, such as brackets, puffballs, earthstars, crusts, spindles and coral fungi, cup fungi, earthtongues, leaf spots, woodwarts, earthstars, and a vast range of microfungi.

Kingdom Fungi is estimated to contain between 2.2 and 3.8 million species, over six times the estimated number of plants, but even after several centuries of study only 144,000 species have been formally described (given an officially recognised name). Currently around 2,000 new species are described per year. The gap between what we think is out there compared to what is known is enormous. In the report ‘State of the World’s Fungi’, the Royal Botanic Gardens Kew estimated that around 93% of fungal diversity on Earth remains to be discovered.
Fungi are an essential part of our natural world, as decomposers and recyclers, engaging in beneficial symbioses with most plants, and keeping plants, insects, and ecosystems diverse and in balance. They are also biodiversity in their own right, and just as important to protect and conserve as other organisms. However, without being to accurately identify fungi it is extremely difficult to either understand what they do, where they are, or how we can try to protect them from habitat loss, pollution, and climate change.
To help improve the identification of fungi, over the past few decades professional mycologists have developed a wide range of DNA-based methods to improve the identification of fungi. The most used method is DNA barcoding – the characterisation and identification of fungal species using sequences of species-specific regions of DNA.
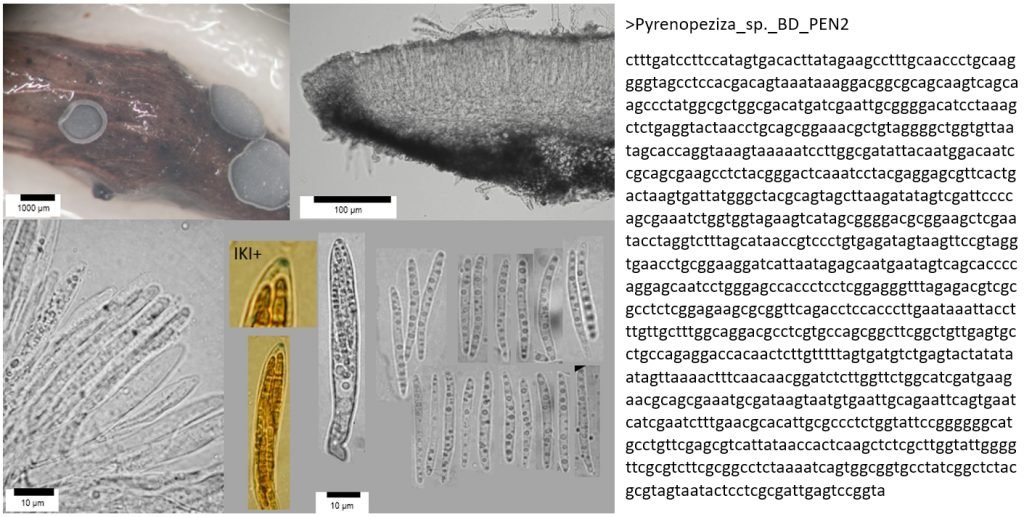
DNA barcoding has become a revolutionary methodology for the identification of fungi, following on from the invention of light microscopy in the 17th Century, scanning electron microscopy in the 1970s, and digital photography and social media in the 21st century. It is morphology-independent, which means it can work hand-in-hand with other identification methods to confirm or refute an identification; be used on its own to compare samples to reference material; and can also detect the same species from soils, plant tissues, water, or air samples. It relies on the assumption that different species have single distinct DNA barcode regions that can be differentiated from those of other species (which isn’t always true using standard barcode regions), but in general is an extremely useful tool for species discovery and identification.
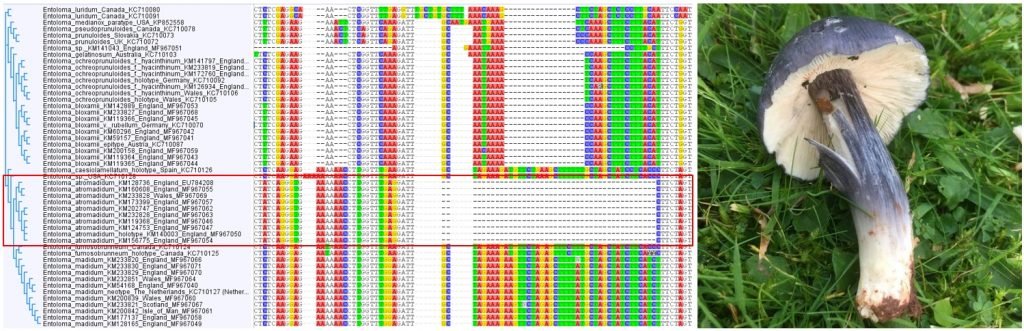
However, a major obstacle to the effectiveness of DNA barcoding has been that it has been limited to a relatively small number of academic scientists across the world, and has been almost inaccessible to the much larger global community of fungus enthusiasts and field mycologists who lack both resources and knowledge to do this work. It has only been in recent years that this scientific divide has begun to be bridged, pioneered by a growing number of “DNA-enabled” members of the public, ranging from youth citizen scientists, to experienced field mycologists in the USA and other countries.
Bento Lab and UK Field Mycologists
In the UK, Bento Lab has been one of the main tools that has allowed UK field mycologists to join this movement.
Pembrokeshire Fungus Recording Network (PFRN)
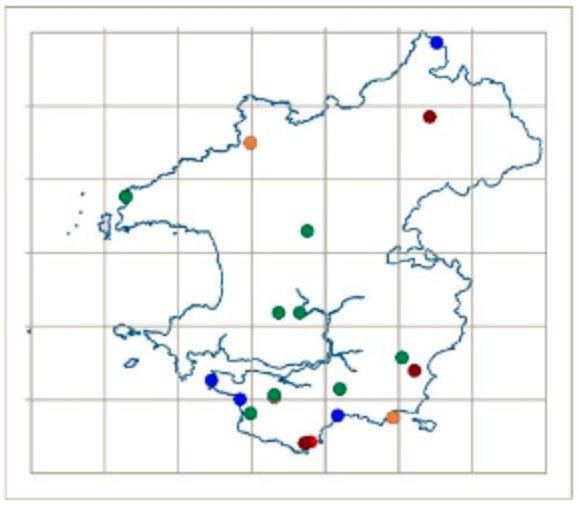
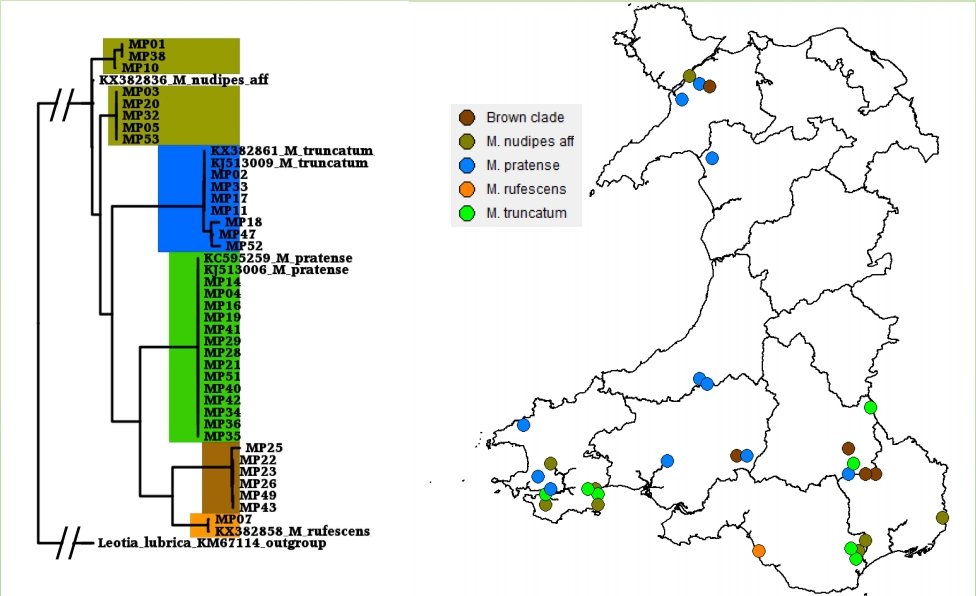
In the UK, David Harries and the Pembrokeshire Fungus Recording Network (PFRN) were one of the first fungus recording groups to become involved in DNA barcoding of fungi. The group initially collaborated with Aberystwyth University Mycology Group in the collection of grassland fungi for barcoding of reference material for soil DNA work. After seeing the DNA barcoding process first-hand, David soon realised that it would be possible for the PFRN to start DNA barcoding specimens if he could source the equipment and reagents.

Members of the PFRN learning DNA barcoding at Aberystwyth University. © David Harries.

After initial work using donated or second-hand equipment, the group became one of the early testers of Bento Lab in 2017, and adapted a quick, safe, and easy “DNA dipstick” extraction protocol for use with fungi. The final step of the laboratory process was the DNA sequencing of PCR products, which was not possible to do at home, but could be arranged at Aberystwyth University. Sequence analysis skills were developed using a range of free software packages and online resources.
Once becoming confident in fungal DNA barcoding, the group began using this methodology to help unravel a number of known cryptic species complexes in Wales, including blackening waxcap mushrooms, and a DNA-based study of green and brown earthtongues in Wales, funded by Natural Resources Wales and the Lost and Found Fungi project (LAFF – see below for details of this project). Ongoing work is addressing some of the groups’ more interesting specimens, including poorly known or potentially undescribed species in habitats important for fungal conservation.


The Lost and Found Fungi project
The Lost and Found Fungi project (July 2014-March 2020) was a citizen science/volunteer orientated project co-ordinated by the Royal Botanic Gardens Kew, funded by the Esmée Fairbairn Foundation. Although the primary drive of the project was volunteer conservation surveying for a defined list of rarely recorded species, it also prioritised enabling and upskilling field mycologists to improve the UK’s capacity for conservation recording and identification. It soon became apparent that a growing number of individuals and groups within the UK fungus recording community, and especially many of the more experienced members, were extremely interested in DNA barcoding their most important finds.
To try to address this need, in late 2018 the LAFF project obtained three Bento Labs and began the process of teaching several fungus groups to use them for fungal ID, adding a further two Bento Labs in late 2019. The LAFF project ran workshops throughout 2019 to introduce DNA barcoding and DNA sequence analysis to interested groups throughout England, Wales, and Scotland; and conducted a joint fungus recording groups DNA workshop in partnership with the British Mycological Society.


A sequencing account was set up with the Core Genomic Facility in Sheffield University to clean and sequence participants’ PCR products using the project’s support budget, and the project provided support for sequence and taxonomic analysis for the generated sequences. Although the LAFF project was only able to provide this support for a limited time before the project ended, by the end of the project in March 2020 there were four fungus groups capable of independent DNA barcoding in the UK, and a further two following close behind. Two of these groups are profiled below.

Sussex Fungus Group
Nick Aplin of the Sussex Fungus Group was one of the first loanees of the LAFF project’s Bento Labs. Although Nick is an amateur expert in field mycology and microscopy he had no previous molecular biology background, but was keen to give the DNA barcoding process a try.

After a few introductory workshops and occasional guidance, plus a lot of independent work while the LAFF project was busy elsewhere, Nick soon picked up the basics and began successfully extracting and amplifying fungal DNA from a wide range of different types of fungi, ranging from large mushrooms to single microfungi fruitbodies less than 0.2 mm in diameter. Notable finds so far include confirming the identity of species of conservation concern; identifying new species of fungi to Sussex, to Europe, and potentially to science; and also matching up asexual and sexual stages of fungi.
Hampshire Fungus Recording Group
The Hampshire Fungus Recording Group is a long-established group populated by very expert and experienced mycologists, regularly making important finds from diverse habitats within the New Forest and surrounding areas. Between them the members possess a wide range of laboratory and technical and computer skills, although none had previously undertaken DNA barcoding work. Starting off with a series of LAFF-run workshops in November 2019, under the lead of Eric Janke the group is now independently producing PCR amplicons and sequences, and has recently confirmed several new species to the UK. Focusing on some of the more difficult fungal groups, they have also been finding previously unsequenced and poorly named species that require further taxonomic work for confident identification.

Amateur fungal DNA barcoding in the UK is still at a very early stage, but successes so far suggest that this work will continue to develop and expand. Several groups are now actively seeking funding to support further sequencing work. The processes of molecular identification can be complicated, but are easily achievable for the amateur mycologist willing to invest enough time into learning molecular techniques.
Hopefully this will help usher in a new wave of UK fungal DNA barcoding and biodiversity discovery, led by amateur experts and fungus enthusiasts rather than just by professionals. After all, it’s much easier to teach experienced field mycologists DNA barcoding methods than teach a molecular biology student how to morphologically identify fungi!
If you’re interested in becoming a field mycologist in the UK, the British Mycological Society has a list of regional groups on their website, and also a very active Facebook Group.
Cover picture: The Urra fieldstation in southern Spain. Credit: Katie Cook
Imagine yourself looking out across a desert landscape, with a warm breeze and the faint sound of busy insects in a dry landscape. This is the setting for the Urra Field Centre in Spain, where this April, Dr Elizabeth Duncan and her colleagues took a group of 40 students for a Mediterranean Ecology field trip.
Based at the University of Leeds, Dr Duncan studies the evolution of bees. There are two different groups: social bees and solitary bees. Social bees, such as honeybees and bumblebees, live in groups and depend on queen bees to reproduce. As their name suggests, solitary bees live on their own. They build burrows underground, or nests in buildings. Solitary bees make up 90% of the bee species in the UK, and bees, together with other pollinating animals, help to pollinate a third of all the food we eat. Like honeybees and bumblebees, solitary bees are under threat from changes to their habitat.

Included on the agenda for the Urra field trip: a project to identify Nosema infections of bees. A fungal parasite, Nosema invades the guts of the bee, and spreads through spores. When infected, bee colonies can fail to grow and dwindle away. For commercial beekeepers, this infection can mean a significant decline in colony productivity. The parasite also poses a risk to solitary bees, as the infection can spill over from commercial colonies into wild bee populations.
To an expert eye, the simplest method to identify a Nosema infection is under the light microscope. However, more accurate tests are possible using genetics. Researchers have developed methods based on PCR, which are sensitive enough to detect low levels of pathogen infection.
Loading the gel to look for evidence of Nosema in our bee samples using the @theBentoLab kit. @e_j_duncan #LeedsMedTrip pic.twitter.com/Dj0chqGLOu
— Chris Hassall (@katatrepsis) April 2, 2019
The sweet sound of science as our @theBentoLab kit whizzes into action, running a PCR on some bee tissue looking for evidence of parasites. Exciting incorporation of molecular biology into the #LeedsMedTrip by @e_j_duncan. Stay tuned for the results! pic.twitter.com/PpbbZfOB5O
— Chris Hassall (@katatrepsis) April 1, 2019
Throughout the week, the team collected honeybees from around Urra as samples for their barcoding experiments. To detect Nosema, the group needed to extract the Nosema fungal DNA, which is only found in the gut of bees. The students dissected the gut of the bees and extracted DNA from a very small portion. Using these samples, they ran conventional PCR assays and visualised the results, all with Bento Lab.
The PCR assay was designed to generate 2 products, the first to confirm the presence of honeybee DNA, and the second to search for the presence of Nosema DNA. In good news for the Urra region, the group found no presence of Nosema DNA in any of the samples they collected.
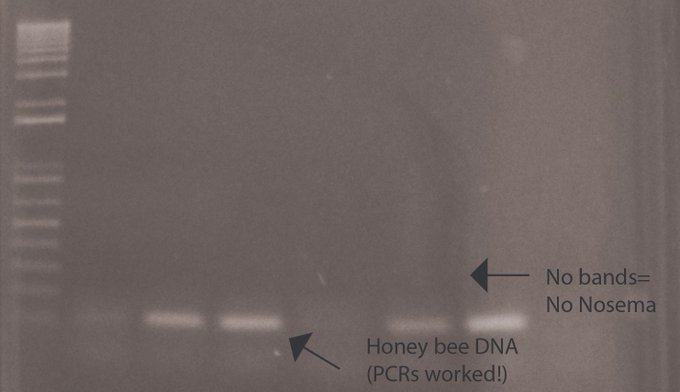
“And the results are in….. – one band for all our samples = Healthy honeybees! No signs of disease yet – good for Urra @katatrepsis @benitez_lab @BillKunin! Now to try more samples & experiments with our BentoLab #LeedsMedTrip” Credit: Elizabeth Duncan
When we spoke, Dr Duncan noted there is limited research available on solitary bees, as they are more challenging to work with. Given the commercial nature of social bees, there is more funding interest in honeybees and bumblebees. It’s also harder to collect wild bees for research, whereas samples of honeybees and bumblebees are easily available from breeders.
In the short-term, there are lots of simple ways that everyone can help wild bees, from growing a patch of wildflowers, to joining a recording scheme to keep track of local bees. If you are interested in supporting wild bees, take a look at Solitary Bee Week to learn more.
I never thought i would be able to do molecular biology in the field @UrraFieldCentre testing wild olive trees, rapeseed and wild rocket @e_j_duncan @katatrepsis , let you know the results later today pic.twitter.com/7UVPeyyjbK
— Benitez-Alfonso lab (@benitez_lab) April 3, 2019
Looking for bee pathogens round 2! #LeedsMedTrip pic.twitter.com/RsLPAeZzsU
— Elizabeth Duncan (@e_j_duncan) April 4, 2019
The field trip this April was a pilot run to test validate the field-based PCR assay. Along with a new group of students, Dr Duncan will be returning next year, and she plans to bring along tools for sequencing onsite for further research, including the portable sequencer MinION from Oxford Nanopore Technologies. Using these state-of-the-art molecular tools, Dr Duncan and future students will expand this work to detect other pathogens, and to measure biodiversity.

We’re excited to announce the release of a new firmware release for Bento Lab. The update brings many new features that have been requested by user feedback, better error diagnostics and critical bug fixes.
We highly recommend all Bento Lab users to update their firmware following these steps. Firmware update can be launched automatically, when Bento Lab is connected to Wifi.
New PCR features
The new software now allows up to 29 different heat stages in a PCR program, with multiple cycles.
Temperature stage timers can now be changed with 5 second increments.
More information is being displayed when a PCR program is running. This includes countdowns for individual stages, and temperature adjustments from touch down.
Saved PCR profiles can be renamed easily from the interface.
Additional changes and fixes
- Error messages give more diagnostic details
- Improvements to touch down performance
- Improved heating and cooling limits
- Updated icons
- Improved Wifi stability
Installation
We highly recommend all Bento Lab users to update their firmware following these steps.
If you encounter any issues during your firmware update, please get in touch with us.
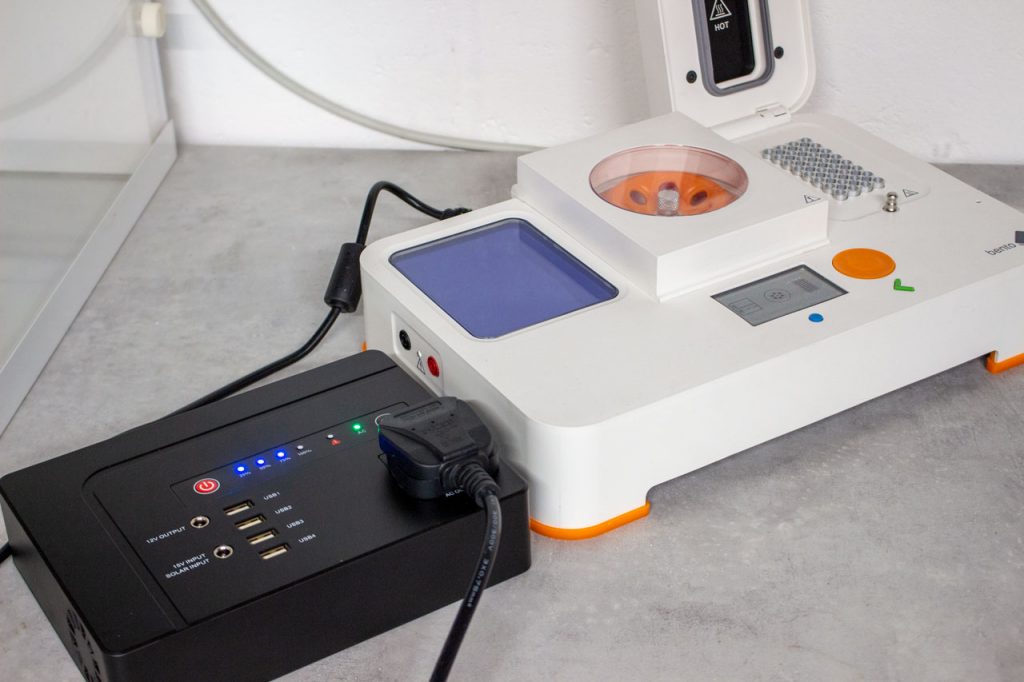
Our users are working with Bento Lab in countries across the world, and Bento Lab comes with a universal power supply. The supply works with all international power systems from Japan (100V) to Europe and India (230V). With the right plug, Bento Lab can be plugged into any standard wall socket to run on mains electricity.
However many of our users are working with Bento Lab in unique environments. To make your research truly portable, you need to keep running experiments when power is not guaranteed. We have been testing out a few different setups to explore what’s possible.
What type of battery can I use?
If you are performing research out in the field, Bento Lab can run off a portable power station. There are two requirements for choosing a power station that will be compatible with Bento Lab.
Socket option
Portable power stations are available with a number of different output/plug options: USB (5V), Cigarette Lighter (12V) and AC mains socket (110-240V). To power Bento Lab, you will need a power station with AC mains plug output.
Power rating
Power stations also have different power ratings, which defines how much energy it can supply in a given moment, for each plug output. As you can imagine, charging an iPhone has a very different power requirement to running a centrifuge. Bento Lab draws a maximum power of 140W. If the power rating is less than 140W, the Bento Lab may not function correctly.
So, to select the appropriate portable power station, you will need a battery with the following features:
- An output with AC mains plug (110V-240V AC)
- A minimum power rating of 140W+
What options are available?
There are a range of power stations available that meet these criteria. We have tested two options: a plane-friendly power station, the Powkey Power Station and a more powerful solar option, the Yeti 400 Power Station.
- Powkey Portable Power Generator has a battery capacity of 146Wh. It weighs 2kg, and comes in a leather carry bag.
- Yeti 400 Portable Power Station can be recharged by solar power, or plugging into a car battery. It has a large battery capacity of 400Wh, and you can add on a solar panel. There is also a smaller version, the Yeti 150.
How much power do I need?
Each module of Bento Lab uses a different amount of energy, and the PCR block consumes the most energy. If you want to travel on planes, we would recommend an AC power supply with a battery capacity of 100-160Wh. If you are traveling by car, a battery with more capacity will allow you to run more protocols before needing to charge.
You can calculate the rough amount of energy used for your protocol. Here’s a breakdown of typical energy consumption for each module over 1 hour.
| Module | Maximum power consumption | Average Energy consumption (60 minutes of use) |
| PCR Thermocycler | 100W (peak 140W) | 100Wh |
| Centrifuge | 15W | 15Wh |
| Transilluminator | 5W | 5Wh |
| Gel electrophoresis | 10W | 10Wh |
To calculate the energy consumed, multiply the fraction of time by the power consumption: 5 min at 30W = 5/60 h x 30W = 2.5 Wh
Example Protocol
Here is a scenario based on a protocol from our community.
Scenario: Prepare a Ready-to-sequence Library
In 55 minutes, Dr Sophie Zaaijer prepared a ready-to-sequence library for MinION, using Bento Lab for DNA extraction. Dr Zaaijer and the rest of the team at the New York Genome Center have developed a rapid, inexpensive, and portable strategy to re-identify human DNA called ’MinION sketching’.
DNA Extraction
- Sample incubation at 56˚C for 10 min. Energy: 1 min x 100W = 1/60 x 100 = 1.7Wh
- Sample Centrifugation. Energy: 10 minutes x 15 W = 1/60 x 15 = 0.3 Wh
Library Prep
- Incubated for 1 min at 30˚C. Energy: 1 min x 100W = 1/60 x 100 = 1.7Wh
- Incubated for 10 min at 30˚C. Energy: 10 min x 100W = 10/60 x 100 = 16.7Wh
Total energy required: 1.7Wh + 0.3Wh + 1.7Wh + 16.7Wh = 20.4Wh
For this protocol, the total energy consumed is 20.4 Wh. This protocol could run 7x on the Powkey Station, or 20x on the Yeti 400 Station.
The koi in your garden pond may be giving the world’s smallest hitchhikers a lift. Over past decades, a number of ornamental fish such as koi and goldfish, have been released into the wild. Either by accident, for example through flooding of ponds, or deliberately, and sometimes illegally, for recreational fishing.
As wild animals, all fish carry parasites. Usually, when imported into another country, fish need to go through a number of health checks. But because they are not for consumption, ornamental fish are excluded from these checks, so they can bring new types of parasites with them that pose a risk to existing populations. When exotic fish escape into the wild, those parasites can end up infecting fish bred for consumption or recreational fishing.
Aquatic consultant, Dr Bernice Brewster is developing new, reliable methods for identifying these parasites using DNA analysis with Bento Lab. Working alongside Kingston University, and the UK Environment Agency, Bernice monitors the introduction of new invasive species, and keeps a record of existing parasites on an electronic database.
“Using DNA to identify these species is fantastic. It’s unlocked a door for me, and it’s creating a whole new scope for confident results.”
During the winter, when farmed fish are less active in the cold, they are moved to new locations to restock fishing lakes. Before moving, the Environmental Agency requires fisheries to conduct a health screening. At this point, a sample of fish are delivered to research professionals like Bernice, and samples of parasites are removed for analysis.
The parasite Dactylogyrus, also known as the gill fluke, is her current research focus. Gill flukes are tiny worm-like animals. They have two pairs of hooks, which the gill flukes use to latch onto their fish host and feed.
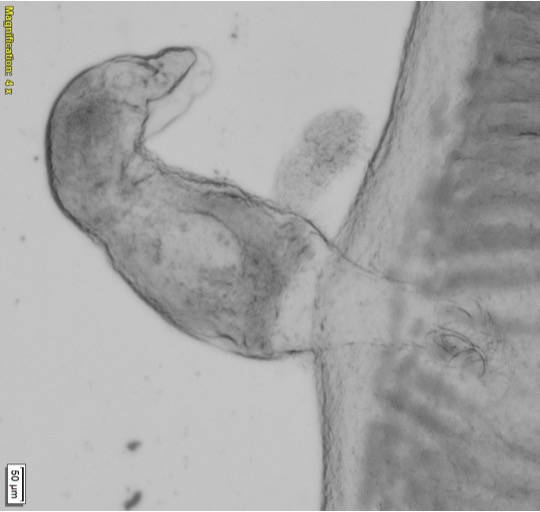
When working with such tiny organisms, size is a big challenge in identifying species correctly. One standard method for identifying gill flukes is light microscopy, where features that are unique to that type of gill fluke are visualised, including the hooks and reproductive organs.
“Gill flukes are only visible at the extreme end of the light microscope.” Bernice explains, “The problem is, these tiny little animals have to orientate themselves in the right position. Even a slight movement can make it very challenging.”

To overcome the challenges of microscopy, Bernice has developed a novel method for DNA identification of gill flukes with Kingston University. Starting by extracting DNA from the samples, Bernice runs a PCR assay, and verifies the product with gel electrophoresis. The PCR product is sent off for sequencing at Dundee University, and once she has the resulting sequence, Bernice runs the code through GenBank to identify the parasite.

As the director of Aquatic Consultancy, Bernice is one of the UK’s many independent biologists, including many working alongside the fishing industry. As a self-employed scientist, Bento Lab is perfectly suited to Bernice’s workflow.
“I am really happy with the Bento Lab. I have 3 PCR programs for my gill flukes. The gel rig is fantastic to check that I have extracted DNA, and I have got a clean sample.”
Since starting the project earlier this year, Bernice and her collaborators have already identified two or three species that have yet to be documented in the UK. Bernice suspects some of these species may even be new to the country, although that is not possible to prove without an existing baseline as a reference. An important outcome of this work is a reliable process to allow Bernice and other researchers to establish a reliable baseline of what species currently exist in the country, which has significant impact of controlling disease in the future.
“In the UK, fish movements are important; controlling what species are moved and where. Establishing a baseline using DNA has the potential to underlie how the authorities control the spread of certain parasites and diseases.”
The E-Gel Electrophoresis System is a convenient, self-contained system for gel electrophoresis. The system uses precast, prestained agarose gels contained within a disposable plastic cassette, which eliminates the need for buffers.
One of our users, Wim Ensink, has made a clever adaptation to enable him to use his Bento Lab with an E-Gel system.
Wim has the E-Gel iBase Power System, an early version of the current E-Gel Simple Runner Electrophoresis Device. He modified a banana plug cable by adding a barrel jack, which allows him to connect the E-Gel system to the Bento Lab electrophoresis output.

Credit: Wim Ensink, University of Amsterdam
“The E-Gel system runs from the Bento Lab. In my opinion, this is really handy, especially for those who have little experience in the lab.”
The E-Gel system is run by a power supply that provides an output of 48V DC, with a current of 0.8A. The Bento Lab supplies an output starting at 50V DC, and Wim noted, “the Bento Lab measures enough current draw at 50V to power the E-Gel”.
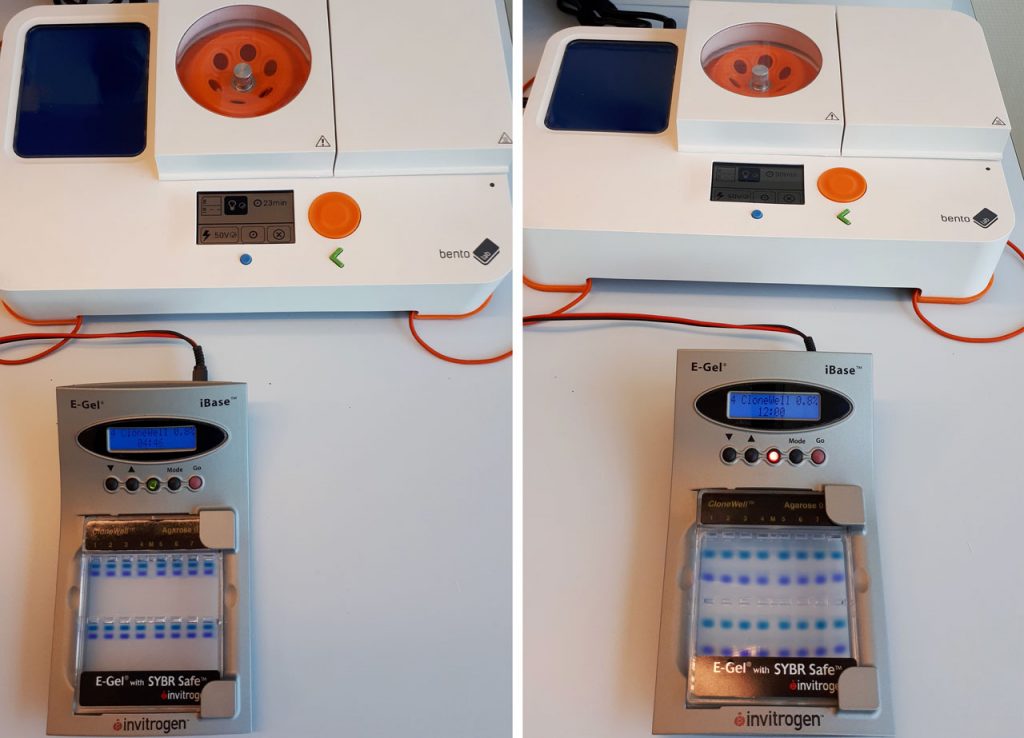
Credit: Wim Ensink, University of Amsterdam
The E-Gel agarose gels are available in a variety of formats, from different stains to different agarose percentages. Wim tested running the set-up with both a 0.8% and 2.0% E-Gel, and both worked well.

Credit: Wim Ensink, University of Amsterdam
The version of E-Gel system that Wim has is a discontinued model, which has been replaced by the E-Gel Simple Runner Electrophoresis Device, and a more expensive option, the E-Gel Power Snap Electrophoresis Device. If you are interested in experimenting, you can also get ahold of older E-Gel systems on eBay from $50+.
Wim thinks it may be possible to make the same adaptions for the current models. “I don’t know if it would also work with the newer type of E-Gel systems, but if the adapter specs are the same, then it probably would.”
For best results with the Bento Lab blue light transilluminator, we would recommend using the E-Gel with SYBR Safe DNA Gel Stain.
Last week, my Bento Lab co-founder Bethan and I spent four days in Norway for the 2019 International Barcode of Life conference (iBOL).

The annual conference focuses on DNA barcoding, a method of identifying species based on sequencing short snippets of DNA. We were invited to exhibit Bento Lab as an exciting option for accessible field research equipment. It was the first science conference we have attended in a while, and we were psyched to talk to everyone about the future of DNA barcoding, and how Bento Lab can contribute.
Check our Youtube channel for a short video blog to get a sense of the vibe of the conference, the pace of conversation at our exhibition stand, and the beauty of Trondheim in the summer.
What is DNA Barcoding?
DNA Barcoding is a method to identify species by analysing a specific region of DNA, the so-called ‘DNA barcode’ region, and comparing it to a reference library. The DNA region depends on the specific biological kingdom. For example, all animals are identified using the same specific DNA region, whilst all plants are identified using a different region.
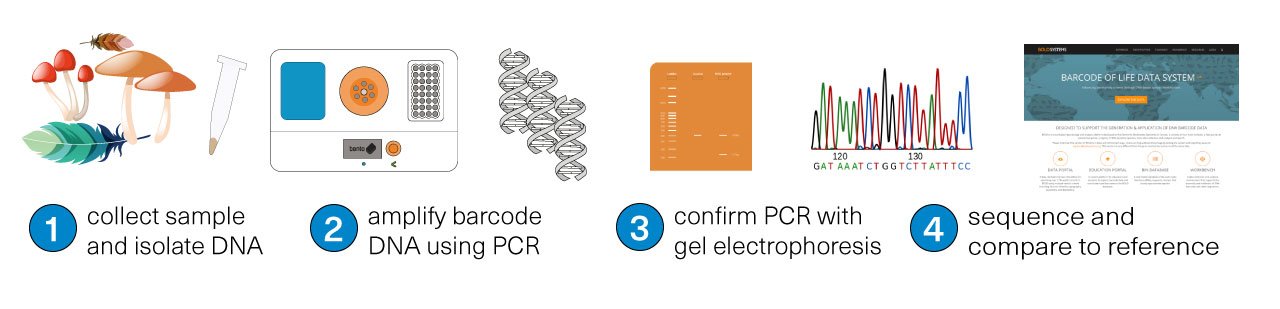
The first step is collecting the sample for study. Then, using a tool like Bento Lab, the DNA is isolated from the sample (1), and the target ‘DNA barcode’ is amplified using PCR (2), meaning that lots of copies of the physical DNA is made. The success of the PCR step can be confirmed using the method of gel electrophoresis (3). Finally, the DNA barcode is sequenced (4). The matching species can be identified by comparing the sequenced DNA barcode to reference databases.
The DNA can be sequenced by sending the amplified DNA to a sequencing service, or by using a portable DNA sequencing machine, like the Nanopore MinION (as some of Bento Lab’s users do) or the Illumina iSeq (like Genome Prairie, one of our communities in Canada).
Tools for field-ready sample preparation

During the four days, we had hundreds of intense conversations about the potential of using Bento Lab for mobile research. The iBOL conference participants are a range of scientists conducting research in a huge variety of settings, so there was a lot of excitement about the mobility and ease of travel with Bento Lab, and we came away with lots of ideas and excitement for the future. There was also a lot of discussion about supporting citizen science groups and school classes to make contributions to DNA barcoding and making a difference to biodiversity. This is also part of Bento Lab’s core mission.
Illuminate Biodiversity to change the way humanity understands our planet
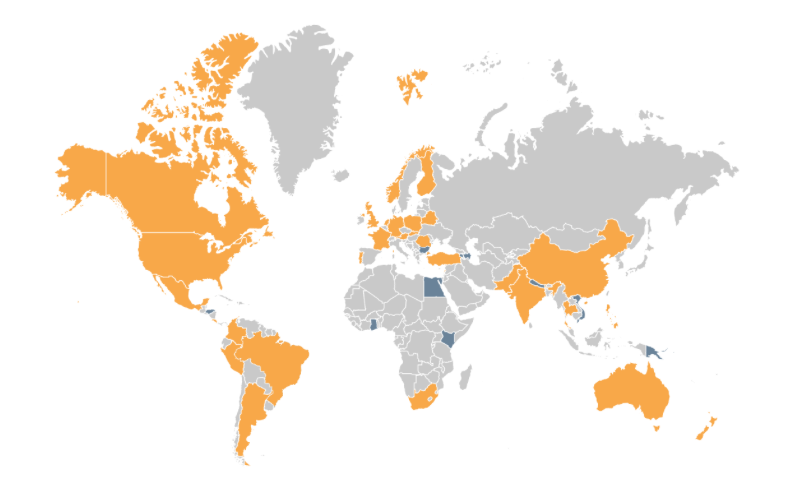
The conference organised by the iBOL consortium, a group of 30 member nations, plus 9 associate nations. All of the nations are supporting DNA barcoding activities in their region to increase the understanding of biodiversity, and shifts in species distribution. Their first major program, BARCODE 500K, finished in 2015 with the successful barcoding of the DNA of 500,000 species. Along the way, this helped demonstrate the value of DNA barcoding at scale, to build “a digital identification system for life”, as iBOL puts it.
The successor program, called BIOSCAN, launched this year. It focuses on three aspects:
- speeding up the discovery of species,
- investigating the interaction between species,
- and tracking shifts in species distribution.
When BIOSCAN is completed in 2025, it is planned to have extended to cover 2.5 million species.
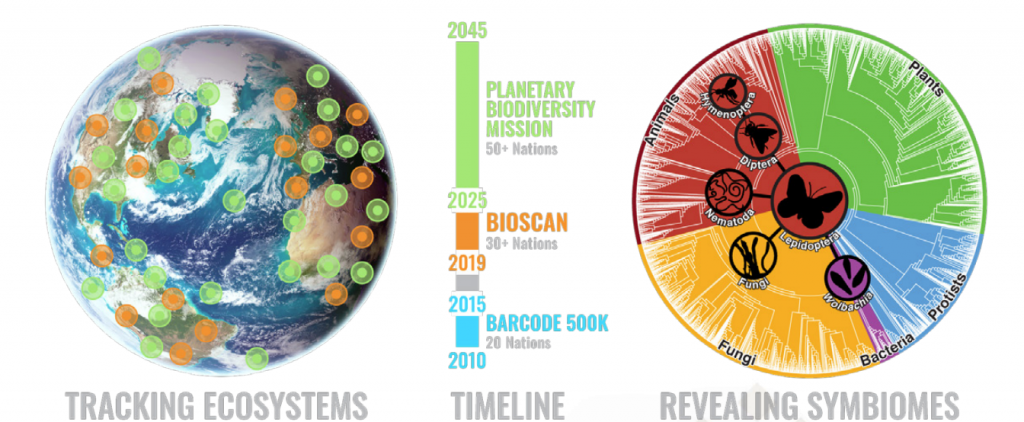
The ultimate scientific goal of iBOL is to build a planet wide biodiversity surveillance system, to deliver a complete and repeatedly updated map of multi-cellular life. This Planetary Biodiversity Mission, or PBM, is targeted to exist by 2045. Ultimately, iBOL is driven by concern for our planet, in the hope that by shining a light on the health of the biodiversity on the planet, we will be able to protect the planet to the benefit of all lifeforms.
Postscript

We had an amazing four days at the iBOL conference. Check out our video blog to get a sense of our time in Trondheim.
I was fortunate to be able to speak to some of the keynote speakers and organisers, including iBOL’s scientific director Paul Hebert, Dirk Steinke and Sujeevan Ratnasingham from the University of Guelph (the “birthplace of DNA barcoding”), as well as Ingrid Ertshus Mathisen and Stine Svalheim Markussen from Artsdatabanken (the Norwegian Biodiversity Information Centre).
Some clips from our conversations made it into the vlog, and we’ll post standalone interviews in the coming weeks. Subscribe to our YouTube channel, so you don’t miss them. I’m very grateful for the time everyone gave me – thank you very much.
Special thanks also to my friend Kazutoshi Tsuda from YCAM who was attending the conference. I have written about his amazing personal biotechnology workshop with Bento Lab and Nanopore MinION sequencing previously.
This month I was fortunate to be invited to Yamaguchi in Japan, to take part in YCAM’s Interlab Camp: Personal Biotechnology.
YCAM, or Yamaguchi Centre for Arts and Media / 山口情報芸術センター, is an amazing media arts center with exhibition spaces, a theatre, workshop spaces, and even a biotechnology lab. Over the course of three days, 30 participants, together with speakers, mentors and the YCAM team, explored the topic of personal biotechnology, conceptually, and in practical hands-on workshops, which also featured the use of 3 Bento Lab devices to prepare DNA samples for Nanopore MinIon sequencing.

YCAM Bio Research
The first time I visited Yamaguchi, which is at the south-west tip of Japan’s main island, was in 2015. I came to YCAM to mentor a synthetic biology workshop as part of the bio-media art group BCL. This workshop was also the first time we tested our earliest working Bento Lab prototypes.

At the time, the team at YCAM was just getting interested in exploring bioscience as part of their media arts practice. Since then, the Bio Research team at YCAM has set up a dedicated wetlab and organised workshops such as Biotechnology from your Kitchen and a series of field trips called DNA of Forests which led to the creation of a botanical guide based on DNA analysis.
Arriving at Interlab Camp: Personal Biotechnology

Interlab Camp was structured around four concepts:
- Reading DNA (first day)
- Writing DNA (second day)
- Bioethics (second day)
- Group work to create conceptual proposals (third day)
The participants included visual artists, high school students, designers, a musician, an anthropologist, and a few reporters. They were complemented by mentors and speakers, including Hideo Iwasaki from Waseda University, Sebastian Cocobia, contemporary dance art group Contact Gonzo, and also myself.

First day: Hands-on DNA analysis
The day started with extracting DNA from different vegetables, which included the use of Bento Lab’s centrifuge. YCAM had set up three Bento Labs shared between the participants.

The lab work was led by YCAM’s Kazutoshi Tsuda, who demonstrated each step. YCAM’s team also had a helper at each table, to make sure no participant was struggling. After the DNA samples were cleaned up, they were set up for PCR amplification using Bento Lab.

While waiting for the Bento Lab units to perform the PCR run, it was my turn to give a talk about the origin, development and ethos on Bento Lab. The talk was going to be live-translated into Japanese, a first for me, which I had been feeling nervous about. Usually I speak quite spontaneously, but this time I had to talk for about two to three sentences, then pause to let the translator take over. However once I got used to it, I quite enjoyed this way of speaking, as it broke the talk into many small chunks, and each time the translator spoke I had a quick pause to consider how to best make the next point.
After my talk and Q&A, the participants resumed the lab work. Using a nanodrop, they measured the amount of DNA that had been amplified. Fortunately, all PCR runs had worked very well, and everyone obtained good results.

For the last activity of the day, each table was given a Oxford Nanopore MinION sequencer. Working in pairs, the participants set up the flow-cells to sequence the DNA they had extracted from vegetables that morning. The first day ended up being a marathon, and we finished after 9pm! The other mentors and I were amazed by the commitment and skill of the participants, many of whom had never handled a micropipette before.
Analyse, Discuss, Conceptualise
The second day and third days built on the “reading DNA” activity of the first day. First, Toshiaki Katayama of Biohackathon showed participants how to analyse the sequencing results of the previous night’s MinION run.

The day continued with talks by Hideo Iwasaki and Sebastian Concobia. Professor Iwasaki is both a scientist (studying cyanobacteria at Waseda University and heading the Laboratory for Molecular Cell Network), and an artist working with biological media. He spoke about his journey as a hybrid artist-and-scientist and his personal home laboratory. I particularly like his piece aPrayer, which takes the Japanese custom of holding memorial services for laboratory animals, and expands the notion to synthetic cells.

Sebastian Concobia, who featured alongside Bento Lab in a Do-It-Yourself Biology feature story of the Japanese newspaper Asahi Shinbun, also gave an inspiring talk about his quest to engineer a true blue rose in his home-based laboratory. The story of a young girl who used his lab to study the nutritional needs of Oxalis Stricta, a plant Sebastian was also studying, stuck with me in particular. She was testing optimal growth conditions of the plant for her school’s science fair, work which resulted in a growth medium for plant tissue culturing of the plant.

On the last day, participants formed groups and worked on conceptual proposals that creatively reflected on the activities and topics of the workshop. After only a few hours of work, each team presented a short talk detailing their proposal. The results included a hilarious presentation on comparing laboratory rituals to the classical Japanese tea ceremony, to probiotic toys designed to strengthen children’s immune systems, to the idea of genetically engineering dogs to be able to drink alcohol – so they can be even better companions.

Genome Bento
A definite highlight of each day was the lunch – a specially commissioned lunch box called Genome Bento.

Bento means “packed lunch” or “lunch box” in Japanese. Each day’s Genome Bento was specifically prepared using only ingredients whose genome had been completely sequenced. On the last day, the lunch was provided as a Genome Bento buffet instead, with ingredients grouped by the year they were sequenced. It was one of the simplest and most effective ways of engaging with the notion of genetic information I have come across so far, and definitely the most delicious.

The first signed Bento Lab
After the project presentations on the final day, YCAM’s Takayuki Ito asked all participants, mentors and staff to leave a mark on one of their Bento Lab units. What a simple and brilliant idea! Doesn’t the final Interlab-Bento-Lab almost look like a rock star guitar? Honestly, I was quite moved by the result. I hope it starts a trend!

All of the workshop’s materials and protocols are on Github.
MakeMagazine Japan has also posted a great write-up in Japanese about Interlab Camp here.
Finally, I would like to thank Takayuki Ito, Kazutoshi Tusda, Fumie Takahara, Kiyoshi Suganuma and Young-Ja Park for inviting and hosting me. I thought the workshop was an amazing achievement and I was honored to play a small part in it.