Key Points
- Bento Lab and MinION enabled in situ DNA barcoding of 32 samples directly in the field.
- The workflow achieved >99.9% accuracy, with 80% of barcodes free from ambiguities – matching lab-quality results.
- All sample prep and sequencing were completed on a boat in under 9 hours, showing true portability for field biodiversity work.
Can you DNA barcode specimens on a diving boat with Bento Lab and MinION, with all of your equipment packed into a backpack? And how many species could you sequence in one batch?
A team of researchers from Singapore undertook the challenge to find out, and produced one of the first and largest reported examples of DNA barcoding with Bento Lab and MinION to date.
Barcoding biodiversity during fieldwork
Portable in-situ DNA barcoding is a promising approach for producing DNA barcode inventories of biodiversity. It can rapidly produce results during fieldwork to help inform further sampling, as well as prevent lengthy delays in obtaining results caused by having to ship and then process samples at researchers’ institutions. However, many current in-situ DNA barcoding approaches are limited to being process only a small number of samples, which can make them less scientifically valuable and less cost effective per sample.
To help solve this problem, a team from the National University of Singapore decided to see how far they could push the limits of in-situ sequencing. Their aim was to develop a workflow to DNA barcode marine animals such as molluscs, sea stars urchins, and sea slugs, with all collecting and processing done during a diving boat day trip. They chose Bento Lab as a portable PCR workstation due to its higher capacity for 32 samples in a single PCR or incubation run.

“We chose the BentoLab over other portable laboratory devices like the miniPCR™ (8-wells) as our proposed workflow was reliant on having a higher thermocycling capacity.” [..] “The thermal-based DNA extraction method complemented the 32-well capacity of the BentoLab and was instrumental in increasing our throughput.”
Chang et al. (2020). Takeaways from Mobile DNA Barcoding with BentoLab and MinION. Genes, 11, 1121. https://doi.org/10.3390/genes11101121
Interested in DNA methods and workflows? Subscribe for monthly insights.
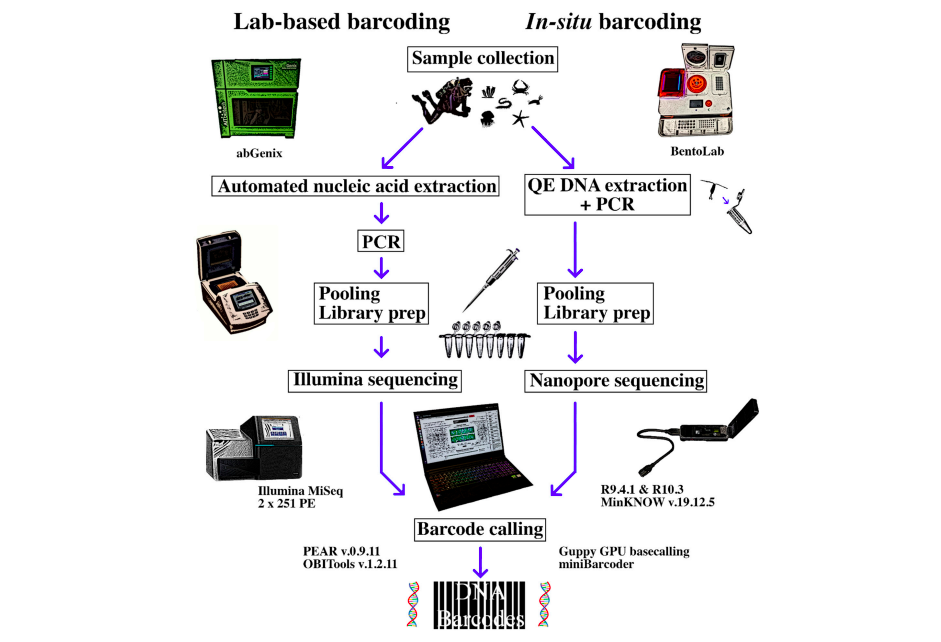
The team of Chang et al. (2020), from the National University of Singapore, put together an entire sequencing system small enough to fit into a backpack, that was designed to have a small footprint when set up on a diving boat with limited space. They built their system around Bento Lab (for its 32 well thermocycler capacity; Oxford Nanopore MinION for sequencing, and a laptop computer.

The on-boat lab setup: a laptop, an Oxford Nanopore MinION, and a Bento Lab. Image CC-BY-4.0 the authors of Chang et al. (2020).
In-Situ Workflow: DNA Extraction, Amplification, and Sequencing
Their workflow had three main requirements: to successfully extract and amplify DNA from a wide range of marine animals; to allow pooling of PCR products into a library for sequencing in a single MinION flow cell; and to allow assignment of reads to samples after sequencing, using a bioinformatics pipeline.
To achieve this, for DNA extraction they chose a one-step enzymatic heat-based method (QuickExtract™ by Lucigen) that would allow processing of 32 samples in under 20 minutes. For amplification they selected degenerate metazoan primers for a short COI barcode region to increase PCR success for a wide range of organisms. To allow quicker PCR product pooling prior to sequencing, and to allow subsequent demultiplexing of reads into samples, they designed unique primers for each sample using 13-bp indexes, thereby removing the need for a separate index ligation step as part of the sequencing workflow.
The researchers wanted to know how successful their quick portable workflow would be compared to what they would normally expect to generate in their home laboratory. So, before their fieldwork, they produced a comparison sequence dataset of 151 samples from 11 phyla collected between 2017-2019 using phenol:chloroform and kit DNA extraction methods, sequencing on the Illumina MiSeq platform.
At the same time, they extracted and amplified DNA from the same samples using their portable workflow to combine with any new field samples. By doing this they could compare success rates between large laboratory and field sequencing datasets, as well as simulate how many samples they could DNA barcode during a much longer field collection expedition.
Once they were happy with the planning of their workflows, the team took a diving vessel on a SCUBA diving trip to collect samples from the Sisters’ Islands Marine Park, Singapore. They collected 31 samples before processing them for barcoding.

Bento Lab was used in the field for:
- DNA extraction, using QuickExtract™ by Lucigen
- DNA amplification, using their degenerate metaozan primers, each sample tagged with a unique barcode
- Oxford Nanopore Technology’s MinION library preparation
The MinION was then used for sequencing for the combined field and laboratory amplicon pools, with the flow cell run for 50 minutes before the team returned home.
The study produced 147 unique DNA barcodes, with 105 produced via MinION sequencing, and 70 sequences that were novel for the existing Singapore barcode database. This represented a substantial increase in known barcoded taxa from the region, achieved quickly and at a relatively low cost.
The researchers reported a field sequencing protocol success rate of around 60% compared to a laboratory success rate of around 77% using phenol;chloroform and kit extractions. Failures appeared to be a result of DNA extraction and PCR rather than sequencing issues, perhaps due to difficult-to-extract tissues or non-optimal primer matching for some species. However, the whole process including new field collections only took nine hours, and any samples that didn’t work could be reattempted when back home.
Based on their results, the researchers concluded that:
- Their “sequencing lab in a backpack” setup could greatly increase sequencing capacity on expeditions or field courses, especially where on-site barcoding is needed but laboratory facilities are lacking.
- Increasing sample numbers was the key to reducing costs per sample, and a quick DNA extraction method combined with Bento Lab’s 32 well thermocycler was critical to achieving this.
- Sequencing accuracy with MinION using an R10.3 flow cell was almost perfect, and in ~80% of samples no ambiguities at all were observed when using a Higher Accuracy base-calling model. Sufficient sequence coverage for all barcodes was obtained in only ~50 minutes, meaning that much of the flow cell capacity was not needed.
Takeaways for Marine Biodiversity Studies and Expeditions
We think that this was an inspiring step forward in field sequencing, especially from a dive boat and with such a wide range of taxonomic groups!
We were also really happy to read that Bento Lab was so useful in increasing sample throughput in the field, and thereby contributing to the knowledge of Singapore’s marine biodiversity. And we hope that the methodology used in this study will be useful to researchers planning similar biodiversity studies in the very near future.
Read the Study
Read the article here: Chang et al. (2020). Takeaways from mobile DNA barcoding with BentoLab and MinION. Genes, 11(10), 1121.
Looking for advice on using Bento Lab?
Book a free consultation or ask a question.