How do you track invisible invaders?
Aquaculture and conservation professionals face a growing challenge: identifying and managing parasites and invasive species that can’t easily be seen or recognized. Traditional tools like microscopy can fall short. But DNA barcoding is making it possible to identify species – even tiny, unknown ones – more accurately than ever before.
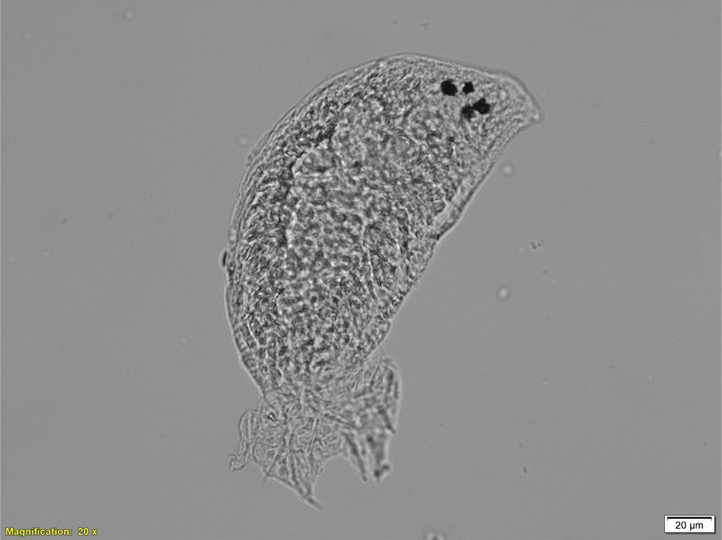
The koi in your garden pond may be giving the world’s smallest hitchhikers a lift. Over past decades, a number of ornamental fish such as koi and goldfish, have been released into the wild. Either by accident, for example through flooding of ponds, or deliberately, and sometimes illegally, for recreational fishing.
As wild animals, all fish carry parasites. Usually, when imported into another country, fish need to go through a number of health checks. But because they are not for consumption, ornamental fish are excluded from these checks, so they can bring new types of parasites with them that pose a risk to existing populations. When exotic fish escape into the wild, those parasites can end up infecting fish bred for consumption or recreational fishing.
Aquatic consultant, Dr Bernice Brewster is developing new, reliable methods for identifying these parasites using DNA analysis with Bento Lab. Working alongside Kingston University, and the UK Environment Agency, Bernice monitors the introduction of new invasive species, and keeps a record of existing parasites on an electronic database.
“Using DNA to identify these species is fantastic. It’s unlocked a door for me, and it’s creating a whole new scope for confident results.”
During the winter, when farmed fish are less active in the cold, they are moved to new locations to restock fishing lakes. Before moving, the Environmental Agency requires fisheries to conduct a health screening. At this point, a sample of fish are delivered to research professionals like Bernice, and samples of parasites are removed for analysis.
Interested in DNA methods and workflows? Subscribe for monthly insights.
The parasite Dactylogyrus, also known as the gill fluke, is her current research focus. Gill flukes are tiny worm-like animals. They have two pairs of hooks, which the gill flukes use to latch onto their fish host and feed.
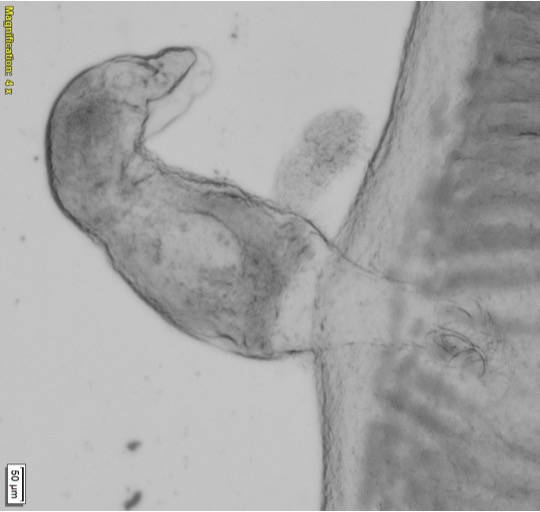
When working with such tiny organisms, size is a big challenge in identifying species correctly. One standard method for identifying gill flukes is light microscopy, where features that are unique to that type of gill fluke are visualised, including the hooks and reproductive organs.
“Gill flukes are only visible at the extreme end of the light microscope.” Bernice explains, “The problem is, these tiny little animals have to orientate themselves in the right position. Even a slight movement can make it very challenging.”

To overcome the challenges of microscopy, Bernice has developed a novel method for DNA identification of gill flukes with Kingston University. Starting by extracting DNA from the samples, Bernice runs a PCR assay, and verifies the product with gel electrophoresis. The PCR product is sent off for sequencing at Dundee University, and once she has the resulting sequence, Bernice runs the code through GenBank to identify the parasite.

As the director of Aquatic Consultancy, Bernice is one of the UK’s many independent biologists, including many working alongside the fishing industry. As a self-employed scientist, Bento Lab is perfectly suited to Bernice’s workflow.
“I am really happy with the Bento Lab. I have 3 PCR programs for my gill flukes. The gel rig is fantastic to check that I have extracted DNA, and I have got a clean sample.”
Looking for advice on using Bento Lab?
Book a free consultation or ask a question.Since starting the project earlier this year, Bernice and her collaborators have already identified two or three species that have yet to be documented in the UK. Bernice suspects some of these species may even be new to the country, although that is not possible to prove without an existing baseline as a reference. An important outcome of this work is a reliable process to allow Bernice and other researchers to establish a reliable baseline of what species currently exist in the country, which has significant impact of controlling disease in the future.
“In the UK, fish movements are important; controlling what species are moved and where. Establishing a baseline using DNA has the potential to underlie how the authorities control the spread of certain parasites and diseases.”