The all-in-one Bento Lab is at least 20-times cheaper than the combination of standalone instruments, and 10-times less heavy, thus allowing for easier manipulation and faster instrument setup. The presented paper provides more evidence of the demand for mobile genetic laboratories.
Mahlerova et al. (2024)
Here’s an interesting portable triplex PCR assay for rapid detection of plant or animal DNA in samples prior to identification using Sanger or third-generation sequencing, which has now been adapted for use with Bento Lab!
Triplex PCR Assay for Animal and Plant DNA
The adapted assay comes from Mahlerova et al. (2024), based on an assay developed by Saskova et al. (2017). The authors’ aim was to produce a fully portable method for applications where the rapid determination of sample origin is essential, for example in the forensic sciences, or to help the interception of protected species of animal and plants during illegal trade transport.
The assay uses three primer sets, two targeting a different taxonomic kingdom (plant or animal), and one producing a larger control band. For plants it targets a 285 bp fragment of the rbcL barcode region; for animals a 511 bp fragment of the CO1 or COX1 barcode region; and it has a 702 bp synthetic internal positive control.
Optimization for Field Use with Bento Lab
To adapt the assay, the authors modified two DNA extraction kit protocols (the Quick-DNA Plant/Seed Kit, and Quick-DNA Miniprep PLus Kit, from Zymo Research) for use with Bento Lab, and validated the assay using Bento Lab for DNA extraction, PCR, agarose gel electrophoresis, and visualisation of the assay results.
They then tested the assay with separate and mixed samples of plant (Hibiscus sp.) and animal (Bos taurus or cow) composition to confirm that both could be detected at once in the same assay.
The results showed that all samples were detected as expected, and this allowed them to send off the PCR products for Sanger sequencing using the same primers as used for the assay. The resulting sequences accurately identified the original species added to the samples.
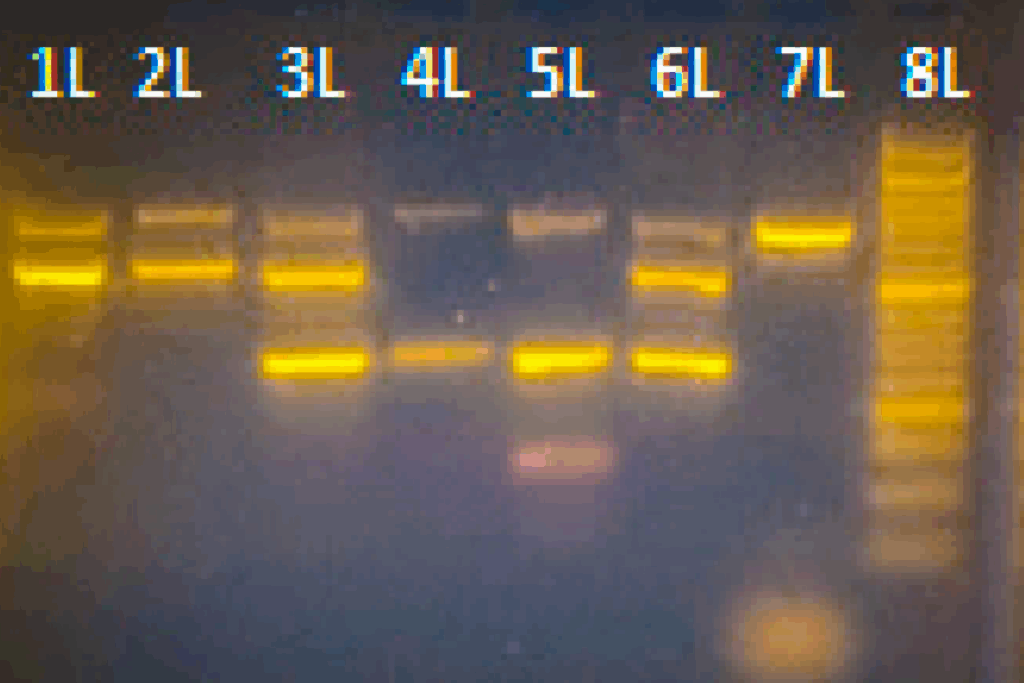
Triplex PCR assay run on Bento Lab
For some samples they observed that high concentration extracts can affect the visibility of the internal control due to the large difference between the plant and internal control fragments. However, this issue can be prevented by dilution of the extract and is a quirk of multiplex PCRs in general rather than being caused by the portable setup.
The authors also discuss some of the restrictions of using a portable PCR workstation such as Bento Lab and how to resolve them, such as the absence of a 2 mL tube heating block, a vortex, and a cell disruptor.
Their approach was to select extraction kits that are easily compatible with Bento Lab’s thermocycler heat block and centrifuge, using plastic pestles for grinding; modified volumes for incubation; and flicking and mixing with a pipette tip rather than vortexing.
However, while the Sanger sequencing used in this assay worked well for mixtures of a single plant and animal, it should be noted that more complex mixtures of multiple plants and animals would require a metabarcoding approach to identify the organisms present.
The authors say that the “simplicity of the Triplex Assay lies in it straight away differentiating between plant/mammalian/mixed sample origin using electrophoresis in a sample of unknown origin, and the fact that single-tube PCR can be used for downstream sequencing analyses”.
We were also very happy to read the authors’ conclusion that “[all] the steps before sequencing can be conducted with the mobile Bento Lab Pro” — great news for anyone aiming to do similar rapid sample identification work in the future!
Read the Study
You can read the article here:
And read the source article for the original PCR assay here:
Saskova et al. (2017). Rapid classification of unknown biological material using a novel triplex assay. Forensic Science International: Genetics Supplement Series, 6, e132-e134 (subscription access only)