Key Points
- To track marine biodiversity, like the presence of box jellyfish or shifting octopus populations, Dr. Ames uses Bento Lab as part of a field-ready eDNA pipeline.
- With passive water sample collection and on-site PCR, researchers can quickly assess species presence and generate actionable data, even in remote areas.
- Dr. Ames trains students from around the world and emphasizes affordability and accessibility. Bento Lab is a key part of teaching DNA techniques that can be used off-grid or in labs with limited resources.
“I went to the Monterey Bay Aquarium where I saw this amazing exhibit of living jellyfish and I knew. “That’s what I’m gonna study!”. It was really love at first sight.”
Dr. Cheryl Ames – Professor, Applied Marine Biology, Tohoku University
Since that first encounter at the aquarium, Cheryl Ames has made box jellyfish the center of her research. The box in box jellyfish comes from their cubic shape, but their geometry is not their most remarkable feature. Box jellyfish are venomous, and their toxins can even kill humans. So her research could literally save your life.
Dr Ames is now a professor at Tohoku University in Japan. Her work is making it possible to predict the presence of specific marine species in different regions of the ocean. Does today look like a cloudy day at the beach, with a high chance of deadly jellyfish? Dr Ames wants to make such predictions a reality.
Detecting DNA in the environment
A key method that Dr Ames uses is Environmental DNA, or eDNA for short. This technique involves analyzing traces of DNA left by species in their environment, for example the water of the ocean.
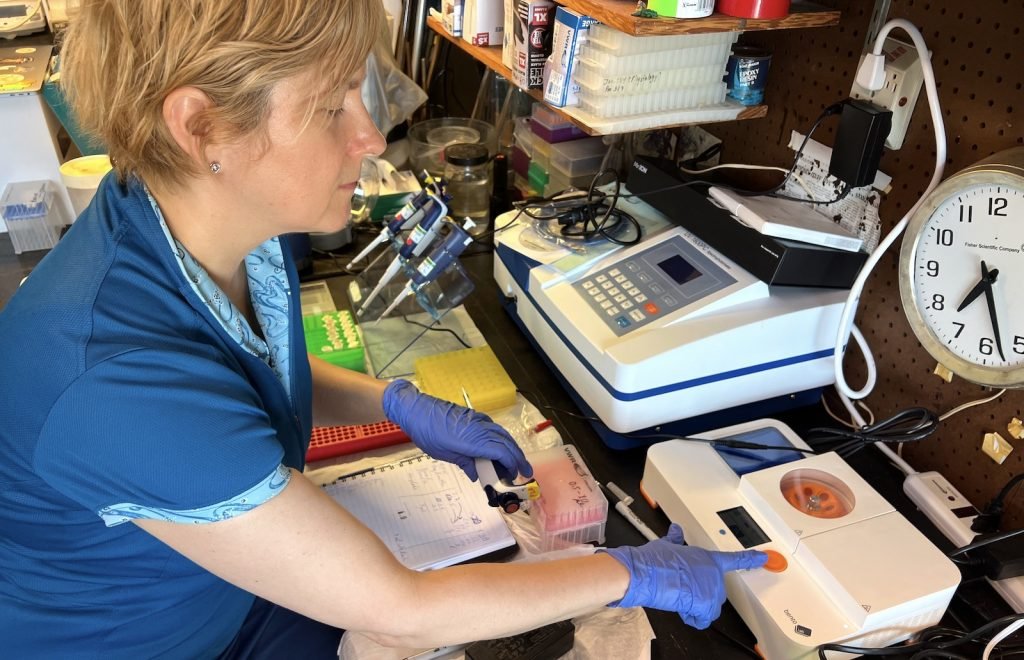
“I think Bento Lab is great for troubleshooting. We can just run the PCRs with 2 or 3 samples and do some quick gels. It’s really handy and we can adjust the settings, temperature, change the primers, etc. I think it’s really a valuable tool for troubleshooting in our lab.”
“I want to be able to detect things with a minimal amount of interference in the environment”, Dr Ames explains. “Of course you need to develop a database, you need to do regular monitoring by taking samples and generating sequences. Right there the Bento Lab is really important for that because you can do the PCRs quickly, and then get the sequences. But you don’t want to go and take every fish in the ocean, right? So eDNA is really important for detecting specific things for which a database already exist or for new discovery.”
Where are all the octopuses going?
The ability to detect species in the ocean with passive eDNA methods creates possibilities beyond protecting beach visitors from deadly jellyfish. For example, fishing and seafood are deeply embedded in Japanese culture, and local fishermen are important community stakeholders for the university. Dr Ames has a background in fisheries and is passionate about the food that comes from the ocean.
“Most of the food in industrialized countries comes from farms. Things that are no longer wild, with thousands of years of domestication. But a lot of the things that we get from the water are still in the wild, so to speak.”
Tohoku University’s campus was badly damaged during the 2011 earthquake and tsunami. Since the disaster, the fishing waters appear to be changing. The number of landings for certain octopus species have been decreasing every year since, while others have increased. So far for the former, fishers have been able to raise the price of octopus to maintain their income, but a tipping point could be looming.
According to Dr Ames, eDNA methods and portable PCR equipment could make it possible to give better advice to the local fishing communities by understanding how and why octopus patterns are shifting, and how to fish sustainably. The key is to make methods rapid, reliable, and field-ready.
“If we know our target, we can bypass metabarcoding methods and just target those 2-3 species that we are interested in”, Dr Ames elaborates. “Then we can go out, take water samples using passive collectors and analyze them. Essentially, all of that can be done on site over a couple of days by bringing a Bento Lab and a battery pack.”


Interested in DNA methods and workflows? Subscribe for monthly insights.
DNA analysis on a dime, where it matters
“I’m always trying to find affordable ways to make science more available to the public and to scientists”
Dr Ames is constantly looking for ways to make DNA analysis workflows more affordable and accessible. The students she trains come not only from Japan, but across the world. She wants to make sure students are empowered to apply their skills even in resource-strapped environments.
“Some of the students might go home and have little access to laboratories and reliable electricity. So if they can learn techniques that they can do back home on low budgets, then this new generation can adapt the methods to their local challenges”.
Ultimately for her research, the distinction between lab-work and field-work is becoming increasingly irrelevant. “I just don’t see the need any more for huge machines to do these things, now that we can run around with Bento Lab and a MinION (Nanopore) and do genomes and metabarcoding on site.”
Top Image by Eugenia Clara
Dr Cheryl L Ames is Professor of International Marine Sciences at Tohoku University Graduate School of Agricultural Science in Sendai Japan. She is a leading expert in marine biology with a research focus on jellyfish systematics, genomics and using eDNA to understand marine biodiversity. She earned her Ph.D. from University of Maryland, funded by a Peter Buck Fellowship from the Smithsonian National Museum of Natural History in Washington, D.C. (United States).
Learn more about Dr Ames’ research and teaching on her website. Recent publications can be found on Research Gate, and you can find her on Twitter @boxjellytalk.
Explore recent publications
- Peterson, M.I. et al. A description of a novel swimming behavior in a dioecious population of Craspedacusta sowerbii, the rediscovery of the elusive Astrohydra japonica and the first genetic analysis of freshwater jellyfish in Japan. Plankton and Benthos Research 17, 231–248 (2022)
- Adriansyah, A.S. et al. Analyses of stalked jellyfish in Kitsunezaki, Japan: Calvadosia nagatensis and two lineages of Haliclystus inabai. Hydrobiology 1, 252–277 (2022)
Looking for advice on using Bento Lab?
Book a free consultation or ask a question.