“The amplification was performed in PPDP with the Bento Lab mobile lab […] to simulate the deployment of mobile diagnostic amenities as a backup to standard benchtop equipment […] or as a deployable option to increase distribution of diagnostic capability in times of crisis.”
Goudoudaki et al., 2021
Researchers from Greece have proposed and tested a rapid and scalable DNA extraction pipeline for emergency microorganism biosurveillance. The pipeline is rapid, inexpensive, and flexible, and it is easily deployable with portable laboratory equipment such as Bento Lab as part of a wider response infrastructure in low resource scenarios.
The researchers suggest that the need for such an approach has become increasingly apparent in recent years, such as when global need to increase Covid-19 testing disrupted normal supply chains and biosurveillance infrastructures. This “wake-up call” suggested the need for protocols and pipelines that are robust in times of crisis.
The key idea was to develop and test a reliable and flexible methodology using readily available reagents that could be integrated with existing infrastructure and adapted depending on local contexts. This would allow essential work to be conducted by anyone, wherever it is needed — whether they are researchers in high-end laboratories, or surveillance dispatch teams set up in improvised laboratories in remote locations using equipment such as Bento Lab.
“This work sought to propose a generic platform that would be adaptable to different needs and applications by the prospective users in emergency settings and with low cost, no royalties or patent and copyrights issues and maximum user-friendliness and sustainability. Thus, the described methodology is not meant to be followed to its detail, but rather to be adopted to local context and application.”
Interested in DNA methods and workflows? Subscribe for monthly insights.
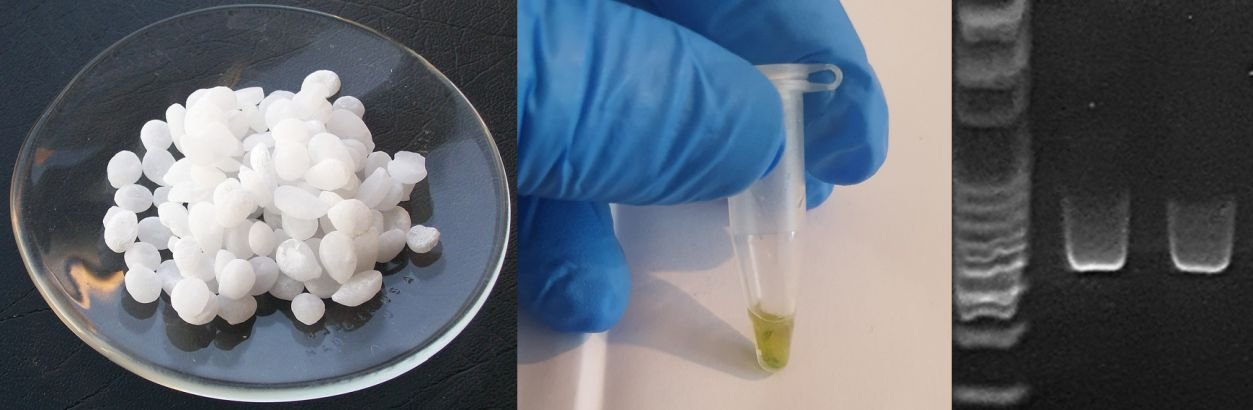
The team of Goudoudaki et al. (2021), a collaboration of researchers from multiple institutions in Greece, developed a DNA extraction pipeline based on strong hot alkaline lysis with a series of optional modifications (see the method at the end of this page). Alkaline lysis is a long-established and popular method for many biological applications, it uses widely available chemicals, and it is rapid, cheap, and easily scalable to large numbers of samples. All of these factors make it an excellent candidate for a generic extraction methodology during emergencies or in resource-limited scenarios.
The team conducted the experimental work using conventional laboratory equipment and Bento Lab to extract DNA, amplify it using PCR, and visualise amplicons using agarose gel electrophoresis. Pure bacteria and fungal cultures were used to represent clinical samples, and plant tissues infected with various agents of disease represented plant pathogens.
The pipeline was tested in two different facilities to simulate its use as a plug-and-play system which could be easily adopted by researchers within a collaborative network. Bento Lab was considered in two roles — a field device for remote settings, and an extension to normal laboratory capacity. The proposed approach meant that in times of need researchers at different institutions, and scientists in the field, could work together without changing their in-house workflows or protocols, but still produce comparable and compatible results.
Overall, the extractions succeeded in producing amplicons for all samples tested for at least one dilution, indicating that this approach can work effectively on a diverse range of organisms and DNA sources. Due to its use of widely available chemicals it was cheap and robust against supply chain issues during times of crisis. The various steps in the pipeline were also rapid, relatively simple to perform, and compatible with Bento Lab’s functionality.
The researchers emphasised that this extraction method is only the beginning model of a generic pipeline. Processes should be continually adapted, improved, and validated based on surveillance requirements and reagent availability. Additional work is also needed in developing approaches for viral threats, and integration with on-site Oxford Nanopore sequencing could make this biosurveillance pipeline even more informative, portable and responsive.
The proposed pipeline is also relevant to much more than biosurveillance at times of crisis. The alkaline lysis method described here, and various variations such as the more dilute HotSHOT method, have been widely used for several decades for many types of biological tissues. It could therefore be a promising DNA extraction method for routine biosurveillance of many different types.
Equally, the distributed model of point-of-need PCR has many applications in non-crisis times, such as in general biodiversity surveying, citizen science, agricultural pathogen testing, education, and many others. The pipeline described here is another important step towards making PCR applications accessible to anyone, anywhere – which is something that we at Bento Lab are very excited about!
Read the open access paper here:
Looking for advice on using Bento Lab?
Book a free consultation or ask a question.Rapid DNA Extraction with Alkaline Lysis (Goudouki et al., 2021)
The alkaline lysis DNA extraction method developed by Goudouki et al. (2021) offers a fast and simple protocol suitable for field-based molecular diagnostics.
Step-by-Step Overview:
- Manual sample crushing in 0.2 M sodium hydroxide (NaOH)
- Incubation at 75 °C for 10 minutes
- Vortexing and centrifugation
- Neutralisation using Tris-EDTA buffer or HCl
Mechanism: How It Works
Under high pH conditions, cells break down, proteins are denatured, and DNA is released into solution. The alkaline environment also inactivates nucleases that could degrade DNA during extraction.
The extracted DNA is generally of sufficient quantity and purity for PCR, especially after 1:10 dilution, and can be used for:
- Target-specific PCR assays
- DNA sequencing for organism identification
Optional Downstream Processing: DNA Purification for Storage
The researchers also proposed downstream processing steps to allow longer-term storage and easy transport of stable DNA to molecular diagnostic laboratories.
Additional steps proposed for stabilising DNA for storage and transport:
- DNA precipitation using isopropanol
- DNA washing and reconstitution in buffer
This method supports a portable, decentralised workflow. It means that sampling, PCR and gel visualisation could be done on site as part of point-of-need sampling, using portable equipment such as Bento Lab, followed by DNA sequencing and identification at regional or national centres.
| Location | Workflow steps |
| On site | Sampling, DNA extraction, PCR, and gel electrophoresis using portable equipment such as Bento Lab |
| Regional or national centres | DNA sequencing and identification |
This makes the method particularly valuable for resource-limited settings and point-of-need diagnostics, and ideal for field use with portable lab equipment.