Hey PCR enthusiasts,
Here are some updates from Bento Lab!
This week we’d love to share details of some useful resources:
- A great modular DNA extraction method (Mu-DNA)
- A free and open-source DNA barcode analysis and organisation platform (BarKeeper)
- A article on problems encountered in DNA barcode practices, that contains one of the best workflow diagrams we’ve seen so far
Methods and Techniques
Mu-DNA: Modular Universal DNA Extraction Methods
If you’re interested in homemade DNA extraction kits, or want to learn how they work, then here is an article for you!
Researchers from the University of Hull (England, UK) have produced, tested, and published a modular DNA extraction system called Mu-DNA (Modular Universal DNA Extraction). Their system is based around spin-column extraction methods using homemade buffers, and it’s flexible enough to extract DNA from a wide range of sample types including water samples, tissue samples, and soil samples.
The authors found their methods produced comparable or better results to commercial kits at a fraction of the cost (£0.67-£0.81/sample compared to £5-£7/sample for a commercial kit). Their modular approach also allowed workflows to be easily optimised, for example by adding different grinding or wash steps for difficult samples.
This article and its protocols are also useful for anyone wanting to learn how spin-column kit DNA extraction methods work because the purpose and function of each chemical is clearly described.
You can find the article here:
You can also read the protocols on Protocols.io.
Software and Applications
BarKeeper, a DNA barcode analysis and organisation platform
If you need a platform to organise and analyse a large amount of sample and DNA barcode data then you might like to learn about the BarKeeper framework.
BarKeeper is a free open-source application developed for the GBOL5 project, a sub-project of the German Barcode of Life initiative aimed at barcoding plant species.
Some key features:
- It keeps track of all your specimen data
- It works for Sanger sequences and high-throughput sequences
- It allows automatic processing from chromatogram to contig to sequence assignment, plus quality checks
- Most if not all of the sequence curation can be done on the platform, including manual checking of chromatograms and contig alignments
- It can be set up on personal computers or on a web server using Docker for easy setup, and can be accessed remotely by web browsers
You can read the article here:
You can also check out the BarKeeper demo portal here: http://barkeeper.uni-muenster.de/ (login: [email protected], password: barkeeper2022).
If you know of any other free, open-source alternatives to managing a lot of DNA barcoding data, then we’d love to know about them too!
Articles We Love
Problems in DNA Barcoding Practices
For anyone interested in DNA barcoding workflows and their problems, or explaining DNA barcoding practices to others, you may find the article below useful.
Cheng et al. (2023) looked at the problems in DNA barcoding in large-scale insect DNA barcoding based on a dataset of nearly 70k barcode sequences. They observed that identification mistakes are not uncommon and can occur at multiple points throughout the DNA barcoding workflow.
To address these problems, the authors emphasised the importance of a detailed quality control procedure that involves checking every sample for mistakes, in which initial potential misidentifications (morphological or genetic) are flagged and assessed, and any mismatches are investigated. This does sound obvious and is what most people do, but it can be difficult to keep track of everything in large DNA barcoding programs involving many people!
We particularly loved their workflow diagram (below) that illustrates the common process of establishing DNA barcodes for specimens (in blue), their quality control processes (in orange), and how the quality control processes are also the steps one might use to identify unknown or suspect samples.
However, it’s important to note that this is an example workflow and your own particular use cases may need additional steps such as re-sequencing, resampling, additional sampling of reference material, phylogenetic analysis, and/or taxonomic work.
You can check out the article here:
If you’ve seen a better diagram that can be used to help explain DNA barcoding in detail to the public and beginners, please do send us a link or reference!
Hey PCR enthusiasts,
Here are the latest updates from Bento Lab!
Bento Lab in the Field
PolBOL, the Polish Barcode of Life project
We were delighted to see the Polish Barcode of Life project using two Bento Labs in their “Biodiversity under your feet” workshop for Tatra National Park, as part of a Biodiversity Genomics Europe citizen science work package.
It’s great to see Bento Labs being used to help promote citizen science and to advance knowledge of biodiversity around the world!
You can check out their post on Twitter/X, and see if you can spot their Bento Labs:
@theBentoLab used in our “Biodiversity under your feet” workshop organised by @POLBOLnode @kzbih_ul for @Tatrzanski_PN within @BioGenEurope citizen science workpackage! Great experience and a lot of fun and interest from the park rangers 😀 Oct 10, 2023
You can find out more about the Polish Barcode of Life project, which is run from the Department of Invertebrate Zoology and Hydrobiology at the University of Lodz, on their website here. Discover more about the majestic Tatra National Park from their Wikipedia page here. And find out more about Biodiversity Genomics Europe here.
Papers We Love:
iDNA — Invertebrate-Derived DNA
Invertebrates such as flies are very efficient collectors of vertebrate DNA from blood, faeces, and dead carcasses. This DNA is called iDNA (invertebrate-derived DNA). iDNA can be used as a convenient way of surveying for vertebrate species using DNA metabarcoding, and to study the community ecology of vertebrates and invertebrates.
A great example of the potential of iDNA is provided by Srivathsan et al. (2022). The authors collected iDNA from flies close to a forest roadside in Singapore; developed a simple and rapid method of extracting iDNA from fly regurgitate and faeces; amplified it using direct PCR; and barcoded it using Illumina and Oxford Nanopore MinION metabarcoding.
The team detected a number of animals from iDNA, including critically endangered species such as the Sunda Pangolin and Raffles’ Banded Langur. By barcoding each fly individually, they were also able to uncover the relationships between fly species and the vertebrate species detected from iDNA. Finally, the team demonstrated that Illumina and portable MinION metabarcoding methods gave similar results, opening up the possibility of this method being even more portable and widely used in the future.
You can read the article here:
You can also watch how their workflow works in this great video on Twitter/X here!
Methods and Techniques
Single-Tube Nested PCR
Nested PCR is a very popular and powerful technique designed to increase the sensitivity and specificity of PCR. It uses two PCRs: an initial PCR with one set of primers, and then a second PCR using the PCR product of the first PCR plus a second set of primers that bind within the previously amplified DNA fragments (i.e. they are “nested” inside the region targeted by the previous primer binding sites). Running two PCRs in succession allows the PCR to be enormously sensitive, while the use of nested primers increases specificity and avoids the non-specific amplification that would usually occur over more than 35 PCR cycles. You can find out more about nested PCR on Wikipedia here.
However, a major disadvantage of nested PCR is that opening up the PCR tubes between the first and second PCRs can increase PCR contamination. This can be a major problem when working with the very low concentration DNA applications that benefit from nested PCR.
We’d like to highlight two very ingenious methods of doing nested PCR that don’t involve any opening of tubes, i.e. single-tube nested PCR. The first method allows you to use any nested primers without designing new ones, but is less convenient to use; while the second method is more convenient to run but may require designing or sourcing some very specific high annealing temperature primers.
Method 1: Spatially separate the primers in the same tube
This method spatially separates the nested primers for the second PCR by drying them onto the lid of the PCR tube beforehand, together with a blue indicator dye (e.g. loading dye), to form a blue spot. After the first PCR, the PCR tubes can be shaken to mix in the second set of primers until the blue spot disappears, and the second PCR can then be run as normal. The primer concentration for the first PCR should be 1/10th or lower of that of the nested primers to ensure that they are mostly used up and don’t compete with the nested primers.
This method comes from Abath et al. (2002), and it allowed the amplification of as little as 1 femtogram of Schistosoma DNA in a single tube, compared to 1 picogram of DNA in their conventional PCR — a 1000x difference in sensitivity! You can read the article here:
A more recent example can be found below, also demonstrating similar success and sensitivity:
Method 2: Thermally separate the primers in the same PCR mix using different annealing temperatures
Design the PCR programs and primers so the annealing temperature of the first PCR and its primers is 10 °C higher than in the second PCR and its nested primers. This means that the nested primers are not able to bind to the DNA in the first PCR due to the high annealing temperature, but are able to bind in the second PCR when the first set of primers are depleted and a lower annealing temperature is used. Again, it is essential to use a very low concentration of primers in the first PCR, e.g. 1/10th of that used in the second nested PCR, to ensure they are depleted in the second PCR.
The method is still commonly used today in many different variations. Below is a great recent study that aimed to optimise this method even further, and to highlight some of the issues that can be encountered with single-tube nested PCR:
If these methods are useful to you, please let us know!
Hey PCR enthusiasts,
Here are some updates from Bento Lab!
Papers we love
Pollen ID using Nanopore-Based Sequencing
For anyone interested in metabarcoding and the study of pollen (palynology), pollinators, and pollinator/plant relationships, here’s a detailed introductory article that takes you step-by-step through pollen metabarcoding using Oxford Nanopore MinION sequencing.
Palynology may seem like a niche topic, but it has many important interdisciplinary applications in allergen monitoring, aerobiology, forensics, honey quality control, paleoecology, plant/pollinator/habitat ecology, and conservation. So we thought it was great to see some guidance on how to make pollen metabarcoding workflows using MinION sequencing.
Also, we’d love to take this kind of workflow into the field with Bento Lab as part of a fully portable “pollen identification” workflow in the future!
You can find the open-access article here:
Methods and Techniques
Booster PCR
Booster PCR is a technique from the early days of PCR which aims to improve the sensitivity of PCR while minimising the production of primer dimers and other artefacts. It’s basically a two-PCR amplification using the same primers for both PCRs, but using a much lower primer concentration in the first PCR to minimise primer dimers and primer-based PCR artefacts.
Booster PCR differs from doing a “double PCR” (using the PCR product of a first PCR as DNA template in a second identical PCR), in that it uses diluted primers in the first PCR. It also differs from nested PCR in that it uses the same primers throughout, rather than a first set of primers and then a set of nested primers.
We’ve written up some notes on this technique and you can read them in our Knowledge Hub here.
We can’t guarantee that Booster PCR will work as well as other methods such as nested PCR, but in some cases it may be more convenient to use. If you think it might be useful to you, why not give it a go?
If it works for you, please let us know!
Methods and Techniques
Touchdown and Stepdown PCR
We’ve also added some notes on our Knowledge Hub about two great methods to make your PCRs highly specific and also give a good yield: “touchdown PCR” and a simpler iteration for basic thermocyclers, “stepdown PCR”. You can find the notes here.
Both methods should be useful to improve the specificity of your PCRs in general, and many of you may already be familiar with both.
Touchdown and stepdown PCR may also be useful in scenarios where you are not sure what is the optimal annealing temperatures for your PCR, for example if you’re using universal primers on a wide range of taxa that might have mismatches to the primers (and hence need different annealing temperatures); or because you’ve modified the PCR in some way to change how the PCR behaves (such as adding MgCl2).
As with Booster PCR, if you weren’t previously using touchdown PCR but are inspired to give it a try, please let us know how you get on!
Hey PCR enthusiasts!
Here are some recent highlights from Bento Lab.
Dipstick DNA Extraction Kits are back in stock
Our Dipstick DNA Extraction Kits have been out of stock for a while, but now they’re back!
For those unfamiliar with it, this kit uses the affinity of DNA for cellulose to extract DNA from samples, crudely clean it, and transfer it directly into PCR mix to act as a template for amplification. It contains filter paper dipsticks, an extraction buffer (a detergent/salt/buffer solution), and a wash buffer (pH buffered sterile distilled water).
The kit is suitable for DNA extraction of tissues from most kinds of organisms (plant, animal, fungal, bacterial), provided they can be manually ground, and it can extract both DNA and RNA. Importantly, it’s quick, easy, and safe to use. However, it’s not a complete DNA extraction method so it won’t be suitable for those wanting to capture all DNA present within a sample, or large quantities of high-quality unfragmented DNA.
The protocol and buffers were originally described by Zou et al. (2020), and you can order the kit from our store.

Interested in DNA methods and workflows? Subscribe for monthly insights.
Methods and Techniques
A guide to MinION DNA barcoding in the field
For anyone considering the possibilities of doing in-situ sequencing with Bento Lab and the portable Oxford Nanopore MinION DNA sequencer, we’d like to share this great literature review by Pomerantz et al. (2022).
The article contains a detailed description of all the steps needed from planning your fieldwork, through DNA extraction, to PCR, library preparation, to downstream analysis. It may be extremely useful for anyone considering this work for the first time, and a good checklist for those with more experience!
The article can be read here: Pomerantz et al. (2022). Rapid in situ identification of biological specimens via DNA amplicon sequencing using miniaturized laboratory equipment. Nature Protocols, 17(6), 1415-1443.
PCR and Education
A PCR assay for onion colour
We were recently asked for ideas for simple and educational PCR assays that could be tried after one had tried all of the assays in our Biotechnology 101 Kit.
One idea we found online was a detailed lab practical outline by Briju & Wyatt (2015), which describes an investigation into the genetic basis of why white onions are white, and why red onions are red. We particularly liked the fact that this practical leads a class though a PCR investigation of several enzymes in onion’s red pigmentation biosynthesis pathway to find out which enzyme is defective, rather than just focusing on one PCR assay.
Here’s the article: Briju & Wyatt (2015). Grocery store genetics: A PCR-based genetics lab that links genotype to phenotype. The American Biology Teacher, 77(3), 211-214.
A PCR assay for rice fragrance
A second idea was a PCR assay developed for rice breeding to determine which strains of rice are particularly fragrant. This experiment could be used to illustrate the genetic basis of consumer-relevant plant traits in a very hands-on way, and you could even test the rice by smell and taste as part of the experiment (outside of the lab of course!). The PCR assay, developed by Bradbury et al. (2005), uses four primers to amplify DNA from a gene encoding betaine aldehyde dehydrogenase 2 (BAD2). In fragrant rice an 8 bp deletion inactivates this gene, allowing the build-up of 2-Acetyl-1-pyrroline, a chemical responsible for the aroma of popcorn, fragrant rice, and fresh baked bread. The PCR assay amplifies a large (577 or 585 bp) amplicon from all rice varieties, while primers specific to the fragrant (deletion) or non-fragrant (no deletion) alleles co-amplify fragments of 257 bp and 355 bp respectively if the alleles are present.
You can find this article here: Bradbury et al. (2005). A perfect marker for fragrance genotyping in rice. Molecular Breeding, 16, 279-283.
If you’re interested in these PCR assays, or have others you are interested in for education or learning, please do get in touch!
Hey PCR enthusiasts!
Here are some recent highlights from Bento Lab.
Bento Lab in the Field
An eDNA suitcase laboratory for WAMBA-NET
We were really happy to see a Twitter/X post by Ouattara Koffi Nouho (@OuattaraKoffiN1) demonstrating his “suitcase eDNA laboratory /DNA barcoding lab in a backpack” (featuring Bento Lab and Oxford Nanopore MinION) to participants of the West African Marine Fish DNA Barcoding Network (WAMBA-NET).
It’s great to see so many participants from different countries coming together to pool knowledge and resources in fish DNA barcoding and eDNA studies to help the study, management, and conservation of West African marine biodiversity!
You can read more about the important and exciting work that WAMBA-NET is doing here.
“Setting up a suitcase eDNA Laboratory/Lab in backpack for barcoding : MinION sequencing”. Wonderful week sharing my experience with WAMBA NET (West African Marine Fish DNA Barcoding Network) participants from Cameroon; CI ; Ghana ; Mauritania ; Nigeria ; Sénégal and France. Sept 19, 2023
Interested in DNA methods and workflows? Subscribe for monthly insights.
Methods and Techniques
If your PCR isn’t working, try diluting your DNA extracts
One simple method we regularly suggest to members of our community who are faced with PCR failures is to try diluting their DNA extracts by 10x to 50x prior to PCR.
It may seem counter-intuitive at first, but PCR inhibitors present in extracts are often the cause of PCR failure. Dilution can reduce PCR inhibitors to non-inhibitory levels, but still leave enough DNA left to act as a template for PCR.
Kemp et al. (2020) provides a good example of how effective DNA extract dilution can be. The authors were trying to amplify DNA from archeological fish bones that were several hundred years old – a very challenging tissue type!
A first PCR pass with undiluted DNA extracts followed by a second pass with 1/10 diluted extracts allowed a large improvement in amplification results. This article also described two other methods that also worked well – “rescue PCR” in which reagent concentrations are increased by 25%, and using a proprietary PCR enhancer called “PEC-P”.
Methods and Techniques
Extract DNA by heating at 98 °C in phosphate-buffered saline
We’re always on the lookout for simple and effective DNA extraction methods to try with Bento Lab, and here is another one for invertebrates, blood, dried blood spots, and feathers (and maybe more?)!
Phosphate-buffered saline (PBS) is a safe, cheap, and easy-to-make salt solution that maintains pH and osmotic pressure (you can find some recipes here). Surprisingly it also appears to be a good buffer in which to heat certain types of tissues to 98 °C to release DNA for PCR.
Jangra & Ghosh (2022) (open access) found this method to be very useful for a wide range of soft-bodied invertebrate species such as aphids, thrips, and whitefly. Squashing a small invertebrate in either PBS and sterile distilled water worked well to produce a crude amplifiable DNA extract, but PBS was more effective in preventing DNA degradation over time.
Thongjued et al (2019) (subscription only) also used PBS heat extractions successfully for a wider range of invertebrates, including insect DNA present in bat pellets. However, their approach did use a PCR inhibitor-resistant polymerase and this may have contributed in part to their sequencing success.
Aslam et al. (2023) (subscription only) reported that heating in PBS is an effective method for extracting DNA from whole blood, blood spots, and feathers for bird DNA applications such as bird sexing. Their method involves a 2–10 min incubation in PBS at 98 °C of a tiny amount of source material, such as using a 10–50x dilution of whole blood in PBS; 1/6th to 2x 6 mm dried blood spot discs in 50 μL PBS; or 2–5 x 2 mm pieces of feather calamus in 20 μL PBS.
Presumably heating to 98 °C in PBS for 10 minutes is sufficient to extract enough DNA from tissues or blood spots for PCR, and denature any proteins that are released, without co-extracting too many other substances that might inhibit PCR.
If you try any of these methods with Bento Lab, do let us know — we’d love to hear how you got on!
Hey PCR enthusiasts!
Here are this week’s highlights from Bento Lab.
Bento Lab in the Field
An Amazing Cave Metagenomics Expedition!
We were delighted to see some first photographs of an amazing expedition into the depths of some of the least accessible, most pristine, and ancient caves in the world, featuring Bento Lab as part of their in-situ Oxford Nanopore MinION sequencing workflow.
The expedition, involving microbiologist Martina Cappelleti (@martycap_micro) of the University of Bolognia, delved into the depths and microbial biodiversity and metagenomics of Imawarì Yeuta, a giant cave system in the Auyan Tepui, Venezuela.
Martina shared a first glimpse of the expedition on Twitter/X:
It was a memorable experience to conduct #insitu #microbiological analyses in the #cave Imawarì Yeuta in venezuelan #tepuis that is one of the less accessible and most #pristine places on #Earth! (1/3) #microbiology #metagenomics #sequencing #nanopore #cave #MinION #exploration Sept 14, 2023
We’re very proud that a Bento Lab was used as part of this ground-breaking expedition, and are looking forward to hearing more about how it went and the results when more details are released!
You can find out more about Martina Cappelleti’s work on her lab group’s website.
Interested in DNA methods and workflows? Subscribe for monthly insights.
Bento Lab spotted at the British Science Festival
This week the Natural History Museum’s Molecular Labs (@NHM_Molecular) ran a stall for the British Science Festival 2023 (7th-10th September), demonstrating how to collect specimens and sequence DNA, and explaining why this work is important.
It was great to see a Bento Lab as part of their demonstration workflow, and we’re very grateful for them tagging us in their tweet so we could see it on display! Check out their Twitter/X post and stand here, and give them a follow to find out more about their work:
We had a busy day yesterday at #BSF23. If you missed us in dome 3, we’ll be back again this afternoon. Find out how to collect samples and extract and sequence the DNA (and why you’d want to!). See our @theBentoLab bento box and have a go on our @brickopore. @BritishSciFest Sep 10, 2023
We love to see Bento Labs “in the wild”, so if you have any photos of Bento Lab to show off on Twitter/X or other social media, please do post them and let us know!
A Free 7-Week Massive Open Online Course on eDNA
Eth Zurich are running a 7-week, 1-2 hour a week Massive Open Online Course on “Environmental DNA: Sensing the Diversity of Life and Assessing Ecosystem Health”, starting on 19th September.
It’s free, and you can upgrade for a small fee to get additional support and a certificate.
You can find the course and sign up here: https://www.edx.org/course/edna-01x
Methods and Techniques
Universal and Rapid Salt-Extraction of High Quality Genomic DNA
If you’re looking for a cheap and safe method of extracting high-quality genomic DNA, that could be used in improvised laboratories or home settings, then this popular and long-established method by Aljanabi & Martinez (1997) may be of interest:
What we like about this method is that despite being 26-years old it appears to work well for many applications — it has 3500 citations according to Google Scholar, and is still in use today! Equally importantly it avoids the use of toxic chemicals, and the chemicals used are fairly accessible with the exception of the proteolytic enzyme Proteinase K.
One nice example of this method in action can be found in Lutz et al. (2023), where the authors found it to be an effective method of extracting high-quality DNA from fish (in this case Southern Red Snapper Lutjanus purpureus).
We don’t currently stock Proteinase K but we would love to know if there is a demand for it, so if this is something you would like us to stock, please get in touch!
Hey PCR enthusiasts!
Here are this week’s highlights from Bento Lab.
Bento Lab in the Field
The curious case of the jelly fungus with snake-like spores

We’re very happy to share this great article by field mycologist Nick Aplin (of Sussex Fungus Group, United Kingdom), on the discovery, characterization, and taxonomy of a tiny obscure jelly fungus, Stypella mirabilis (now Myxarium mirabile), DNA barcoded using Bento Lab 🙌.
Tiny jelly fungi are often more poorly investigated than the larger fungi. In this particular case, Nick was surprised and intrigued by the unusual snake-like spores of his collection. After DNA barcoding it using Bento Lab and the filter-paper DNA extraction method, the sequence didn’t match anything well in DNA sequence databases. But he suspected it could be a described species that hadn’t yet been sequenced.
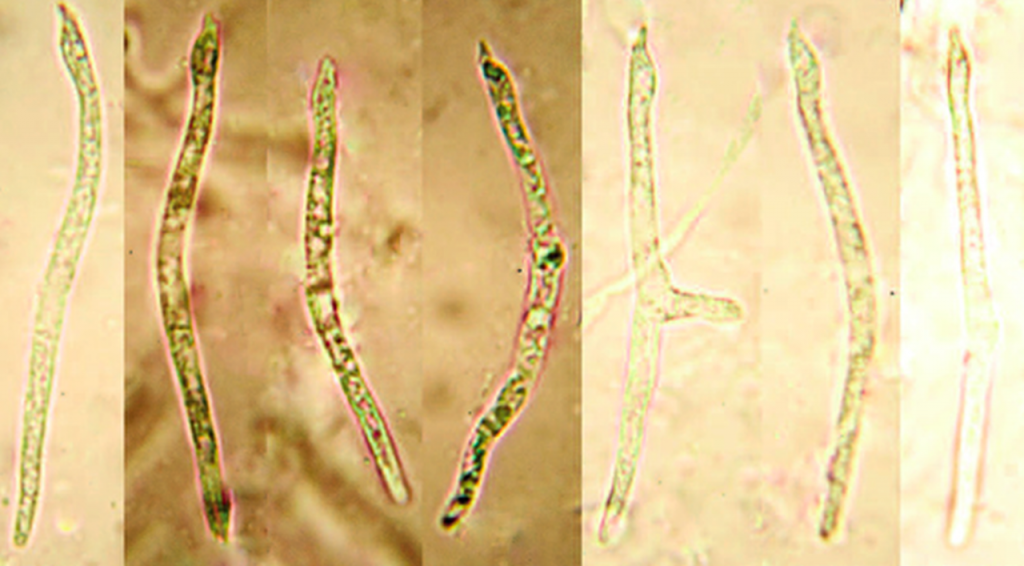
After correspondence with Dr. Peter Roberts, a world expert on the jelly fungi, it turned out that this collection was identical to a species he had described back in 1998, Stypella mirabilis. The epithet “mirabilis” comes from the Latin “to marvel at” and “wonderful”, reflecting the fact that he was also surprised and delighted by the spores. It was also in his opinion a very rare species, a suspicion supported by the lack of UK or global records between its description and Nick’s rediscovery. The DNA barcode indicated that it now would be better placed taxonomically in the genus Myxarium (along with many other species previously placed in Stypella), so Nick formally proposed the name of “Myxarium mirabile”,
Nick’s article expanded the knowledge of this extremely rarely seen fungus; provided a first DNA barcode; and improved its taxonomic classification.
Amazing work for someone doing the work for fun and personal scientific interest, with a microscope and Bento Lab in a spare room!
You can read the open access article here.
Interested in DNA methods and workflows? Subscribe for monthly insights.
Accessible PCR Spotlight
Identifying animals using DNA from their scat
We really love the idea of portable PCR applications becoming useful tools for ecologists and citizen scientists in surveying, monitoring, and identifying biodiversity. DNA barcoding is one application which many people find valuable for discovering and recording biodiversity, such as fungal barcoding. But what about other applications, such as specific PCR assays for particular animals?
We took a look at the feasibility of using Bento Lab to identify bats from their droppings, and thanks to some amazing publications by researchers at North Carolina University and elsewhere we think this would be very achievable, either with standard barcoding or with Oxford Nanopore MinION metagenomics. You can read our thoughts on this, with links to their articles, here.
Specific DNA assays for different target species would also be very possible, and we chanced upon a very nice article by Wilkinson et al. (2022) that used a workflow that allowed identification of arctic and red foxes from their scat using agarose gel electrophoresis. Their workflow could also broadly identify what they had been eating! You can read this article here. We would love to explore this kind of application further if anyone is interested in doing it with Bento Lab!
Paper Highlight
A new strategy for sexing parrots by Kroczak et al. (2021)
Parrots are an important group for veterinarians, bird breeders and conservationists, and knowing their sex is important for a range of bird health, breeding, and ecological research reasons. But they are often difficult to sex without using DNA, especially when very young. Unfortunately, parrots are quite genetically diverse. There is no single primer set that allows reliable sexing for all species, and information on which primers work best with specific parrot species is sparse and scattered in the literature.
We recently wrote a short note on a great article by Kroczak et al. (2021) who developed a new strategy for sexing the order Psittaciformes, using multiple primer sets and a strategy of using the most generally successful primers first to confirm results. The authors reported the results of these primers on male and female birds of 135 species of Psittaciformes, so users of the strategy will know exactly what to expect in terms of banding patterns and amplicon sizes.
You can find our note on the article here.
We don’t yet stock these primers (…yet? Let us know if you would like them). But we would be interested in hearing if there was any demand for these from any PCR enthusiasts or bird breeders!
Hey PCR enthusiasts!
Here are this week’s highlights from Bento Lab.
Bento Lab Upgrades
- Software Update
We have released a new version of the Bento Lab firmware. Based on your feedback, we have cleaned up some confusing interfaces. The update also fixes a bug when changing the centrifuge run timer and other small improvements.
You can follow this guide to install it on your Bento Lab. - Hardware Upgrade – A better PCR lid latch
We are always making small improvements to Bento Lab. In the past weeks we have been testing a new PCR lid latch. By switching this small part to a new elastic material (and going through many cycles of fine tuning) we have been able to make the PCR lid opening and closing experience much more robust and consistent.
If you want to upgrade your existing Bento Lab lid latch, you can order a free replacement part on our website here.
Replacing the latch is a simple procedure that takes less than 5 minutes, and requires only a basic screw driver set.
Interested in DNA methods and workflows? Subscribe for monthly insights.
Bento Lab on a Boat, featured by Oxford Nanopore
We were excited to find that the DNA Barcoding on a Boat story we shared with you two weeks ago was featured by Oxford Nanopore on Twitter (or “X”).
If you have been using Bento Lab and ONT as part of field sequencing workflows, we’d love to hear from you. It would really make our day!
DNA barcoding can be used to identify organisms, but in most cases requires samples to be sent to a lab and are limited to a small number of samples. In this blog, learn how the MinION can be used anywhere to rapidly obtain results in less than an hour:
— Oxford Nanopore (@nanopore) Aug 30, 2023
Inspiration for Educators
Finally, we saw this fun video from Florian Leese of a hands-on eDNA teaching day and wanted to share it with you. Integrating real research into teaching takes effort, but it pays off, and this is a great example.
Intense Day2 of #MolEcol23 module ended successfully. 7 hrs sampling of sites along the #Kinzig river. Then students processed their samples & started (e)DNA extraction 🙏👍🏼 Much longer than a regular teaching day – but that can happen if one includes real research in teaching.
— Florian Leese (@leeselab) Aug 29, 2023
Hi PCR enthusiasts!
Here are this week’s highlights from Bento Lab.
Bento Lab in the field
Bento Lab+ONT for clinical diagnoses in resource-limited scenarios
Could Bento Lab and Oxford Nanopore Technologies MinION sequencing be a solution to lack of medical molecular labs in resource-limited countries?
A team of researchers involved in the SeqTanzania project, an initiative aiming to decentralise sequencing and bring it to wherever it needs to go in Tanzania, believe it could be.
Kumburu et al. (2023) published a case study using a Bento Lab and ONT MinION to rapidly investigate potential causes of a serious and acute diarrhoea in small child, where they identified the causal agent as the bacterium Campylobacter jejuni within 24 hrs. Following this diagnosis the child was prescribed erythromycin (an antibiotic commonly used to treat Campylobacter), had almost immediate improvement, and was discharged several days later.
Although similar real-time diagnostics have been published before in the literature, this case appears to be (to the best of the authors’ and our knowledge) the first description of a completely field-deployable sequencing set up, involving all steps from DNA purification, sequencing, and bioinformatics analysis, on a real-time patient sample for clinical diagnostics. If you know otherwise, please let us know.
The most remarkable part of this study is that the researchers were in fact on their way to a field site to test environmental water samples when they received the call, before rushing to the hospital to undertake a completely different kind of metagenomics!
Here’s the article:
Interested in DNA methods and workflows? Subscribe for monthly insights.
Ideas for Educators
DNA Barcoding Education Projects
For anyone interested in helping to raise the next generation of biologists, ecologists, bioinformaticians, and taxonomists, we’d like to share a really inspiring article by Henter et al. (2016).
The article looks at five different DNA education projects, and discusses how DNA barcoding can be used in a range of different education contexts for students, teachers, research projects, and environmental barcoding surveys.
Thousands of DNA barcode sequences were produced by the various participants and a very large number of people were engaged and learned more about DNA. Importantly, the five projects produced a lot of valuable insights that could be useful for anyone planning future projects in DNA barcoding and education.
You can read the article here.
Tips and Advice
Using Bento Lab without a microwave during fieldwork
Some of you have asked us “can we use Bento Lab without using a microwave oven to melt agarose for electrophoresis gels during fieldwork?”
So we wrote an article describing the pros and cons of several approaches, such as:
- Using a rice cooker or other small heating device
- Using a camping stove
- Using your own pre-cast gels
- Using E-gels
Some of you (many thanks 🤗) have also sent us feedback and suggestions via Twitter and LinkedIn, so we have added these improvements to the article.
Read more here – and if you are planning a field trip or have cast gels in the field, we would love to hear your thoughts and suggestions.

Pouring agarose from a traditional Turkish coffee pot – a suggestion made by Ineke Knot.
Image © Ineke Knot
Looking for advice on using Bento Lab?
Book a free consultation or ask a question.Hi PCR enthusiasts!
Here are this week’s highlights from Bento Lab.
Bento Lab in the field
DNA barcoding on a dive boat
We featured a really nice example of in-situ DNA barcoding during fieldwork, in which a team of researchers from the National University of Singapore developed a DNA barcoding workflow for marine animals (from worms and sea slugs to fish) that could be packed in a backpack and operated on a diving boat during a day trip.
The authors chose Bento Lab because its 32 well thermocycler would maximise sample processing capacity. They combined it with a quick DNA extraction method, tagged barcoding primers to allow sample demultiplexing, Oxford Nanopore MinION for sequencing, and a laptop. Because everything fit into a single backpack, it could be used as a small sequencing workstation in the afternoon after SCUBA diving for samples.
It’s a really great example of what can be done with portable equipment such as Bento Lab and MinION, and with some clever workflow design to maximise sequencing capacity and speed! The workflow is also a very good resource for anyone planning similar field DNA barcoding work with Bento Lab and MinION, whether on boats or on dry land.
Read our story on their article here.

Interested in DNA methods and workflows? Subscribe for monthly insights.
Tip for Educators:
Free Genomics for Educators Online Course
Some of you will be interested in Genomics for Educators, a free online course by Wellcome Connecting Science. It is designed to help teachers and educators learn strategies and techniques for teaching genomics in and outside of the classroom to create enriching learning opportunities.
Genomics for Educators starts next week on Monday 21 August. It runs over three weeks, and you can complete the content at your own pace. The course is accredited by the Royal Society of Biology and CPD UK.
You can find out more and sign up here
Methods and Techniques:
“Dust” Metabarcoding
We spotted an article from 2017 speculating that, in the future, equipment such as Bento Lab, MinION, and robot vacuum cleaners could be used for citizen/community science metabarcoding of the dust microbiome.
Metabarcoding is a method by which DNA barcode regions of all the organisms in a sample are extracted and sequenced all together, to allow identification of all the organisms that have contributed DNA to that sample. So it’s an amazingly powerful method of using DNA to look at the world around us. Environmental DNA (eDNA) surveys often use metabarcoding methods.
The Bento Lab/MinION/robot vacuum cleaner workflow hasn’t turned into reality yet, but we do love the idea! And it still could be possible – research using DNA metabarcoding of dust or particles in the air is becoming an increasingly well developed methodology, and is producing a lot of exciting research.
For example, researchers are metabarcoding plants from airborne dust traps for biodiversity surveys, for forensics, and to link respiratory allergies and asthma cases to particular grass species. Animals can also be metabarcoded, for example to detect illegal wildlife trade in sharks and rays, or to detect animals within a large site over the course of a year. The sky is the limit — literally so if you attach air samples to an aeroplane and then metabarcode collected particles.
So there is enormous potential for metabarcoding for all sorts of applications. We’re very excited to see how Bento Lab could be used to help make this kind of work more accessible and portable in the future.
Looking for advice on using Bento Lab?
Book a free consultation or ask a question.