Overview
W and Z bird sex chromosomes
This experiment targets the CHD1 (Chromodomain Helicase DNA Binding Protein 1) genes, which are present on the W and Z sex chromosomes of birds as two variants – CHD1-W (on the female-specific W chromosome) and CHD1-Z (present in males and females on the Z chromosome). These genes are homologous (almost identical in structure and function) but contain introns (DNA that is removed by RNA splicing before it is translated into mature RNA) that usually differ in length between sexes within a species and between species.
What are the possible results for this experiment?
There are two possible results for most birds:
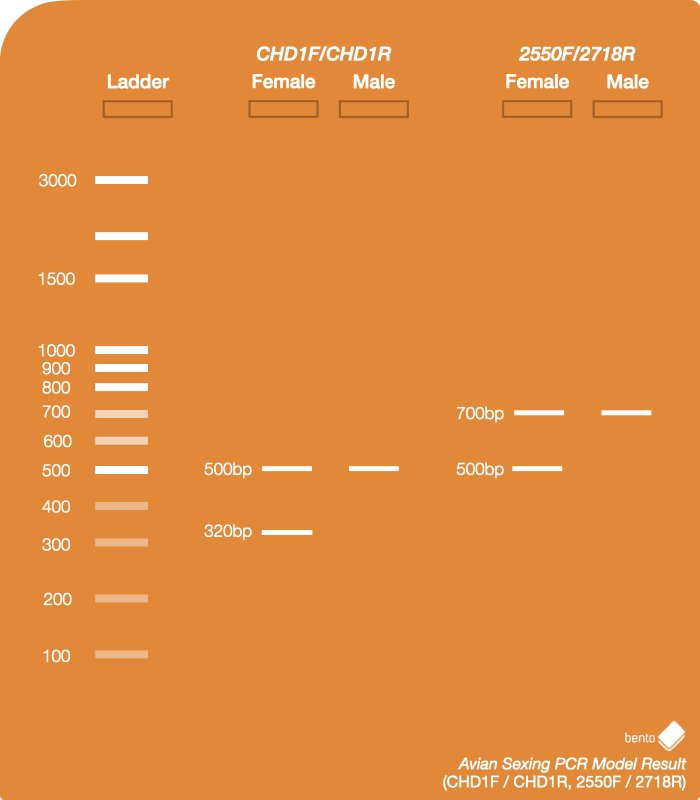
Female: One copy of the CHD1-Z gene is present on the Z chromosome, and one copy of the CHD1-W gene is present on the W chromosome. The target amplified regions of these genes are generally different in length and will be visible on an electrophoresis gel as two distinct bands.
Male: Two copies of the CHD1-Z gene are present – one on each of the Z chromosomes. The amplified regions of these genes are almost always identical in length and will be visible on an electrophoresis gel as a single band.
Exceptions to these results may occur and are detailed below.
False positives (detecting males as females)
a) Contamination from previously amplified female bird PCR products on pipettors, solutions, workspaces, tubes, or pen, may result in false positives. Any amplicons produced by the negative control are a sign of potential contamination. Good PCR lab practice should be undertaken to prevent contamination. See the “Good PCR lab practice” resource for more information.
b) Very rarely a male bird may possess two different variants of the CHD1-Z gene in its Z chromosomes (Z allele polymorphism) that can show as two bands (Dawson et al. 2001). Even more rarely two different variants can form a heteroduplex molecule (two mismatched single strands of DNA bound together in a double stranded molecule), resulting in a hybrid molecule that migrates more slowly through the gel than would be expected (Casey et al. 2009).
False negatives (detecting females as males)
a) Contamination from previously amplified PCR products from male bird samples may result in false negatives (see the section on false positives above).
b) Some bird species may have very similar sized introns in the target CHD1-Z and CHD1-W gene regions. Samples from these birds therefore produce two very similar sized amplicons for both regions, which may be visible as a single band even if the bird is female.
c) The primers may not be a good match for one of the target gene regions in females, e.g. in some Passerine birds using the 2550F/2718R primers (Fridolfsson & Ellegren 1999), or some Psittaciformes using the CHD1F/CHD1R primers (Çakmak et al. 2017).
d) Shorter amplicons may be preferentially amplified, resulting in only a single band being visible when run on an electrophoresis gel. However, it may still be possible to identify single band results as female if the size of expected amplicons is known when using CHD1F/CHD1R and 2550F/2718R primers, since the female specific female-specific CHD1-W region is smaller than the shared CHD1-Z region.
Because of the potential for false negatives and positive results (particularly females being identified as males), we recommend investigating the published literature to determine if anyone has previously used the CHD1 region to determine the sex of the bird species of interest; what primers worked best for that species; and the expected amplicon lengths for males and females.