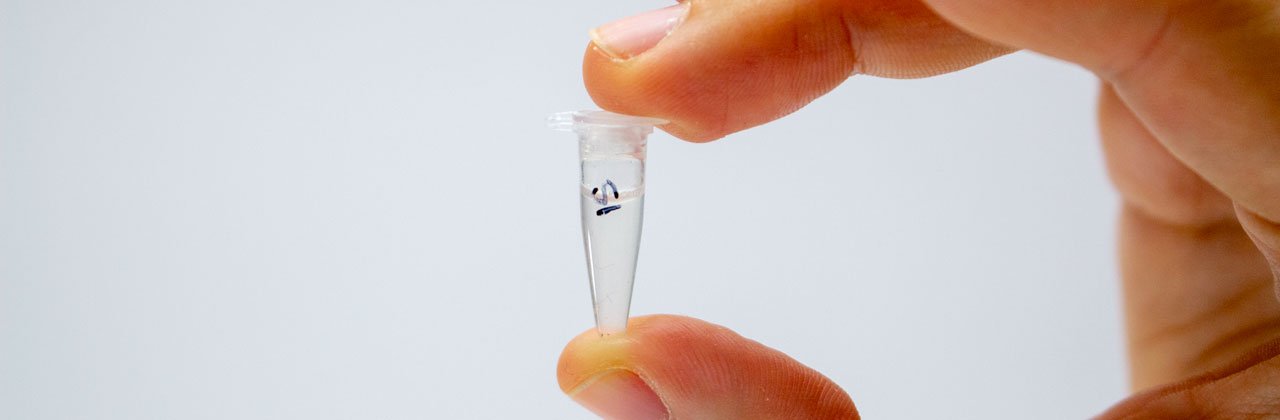
Collect a sample from the fungus or mammal you would like to DNA barcode.
- For macroscopic fungi (mushrooms), break off a small section from the edge – no need to take the entire fruiting body.
- For microscopic fungi, use a sterile swab to collect mycelia from the culture.
- For live mammals, obtain a spit or blood sample.
- For meat products, use fresh or remove a chunk from a frozen sample and allow to defrost.
- For plants, rip off a small section of leaf.
Use a new scalpel/razor blade on a clean surface (e.g. a clean piece of paper towel on top of cutting mat/non-scratch surface) to prepare a very small section of sample approximately 5 mm3 diameter. At this stage, if possible, try to shave off surfaces of the sample that have been exposed as these could be contaminated with microorganisms.
To avoid sample cross-contamination, sterilise or change gloves and scalpel/razor blade between samples and use a new piece of paper towel each time.
For fungi and plants, you can instead use a pestle and mortar to pulverise your sample and then scrape 5 mm3 of pulp into the PCR tube.
Place the sample in a PCR tube labelled on the top and side in permanent marker.
Even if you only have one sample, it’s good practice to label the tube clearly with a unique identifier. It’s also a good idea to mark on the tube the date and to keep a note somewhere of which samples were prepared, and when.