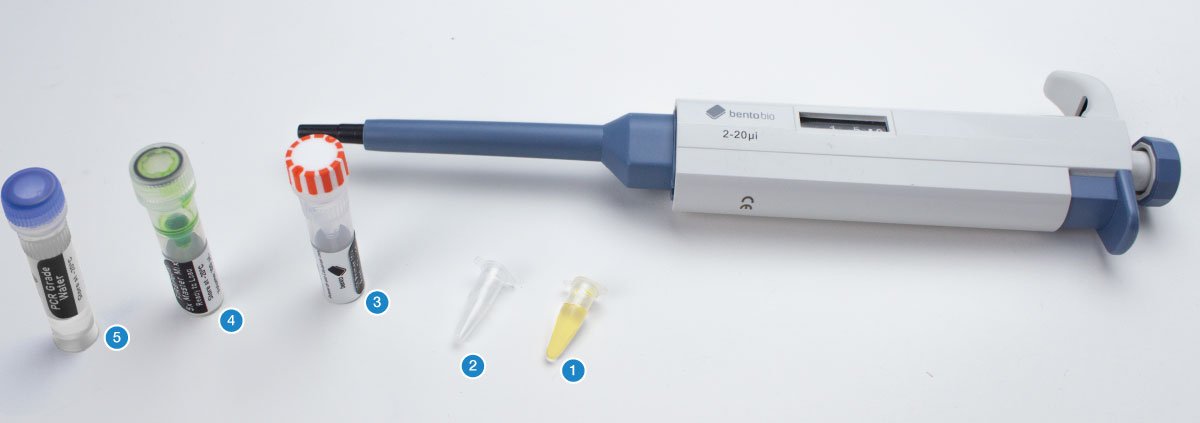
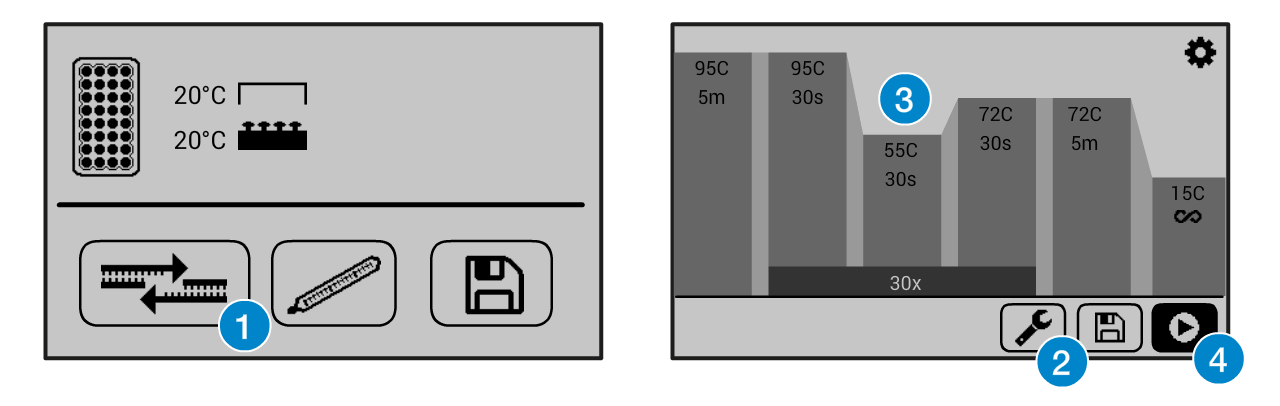
Overview
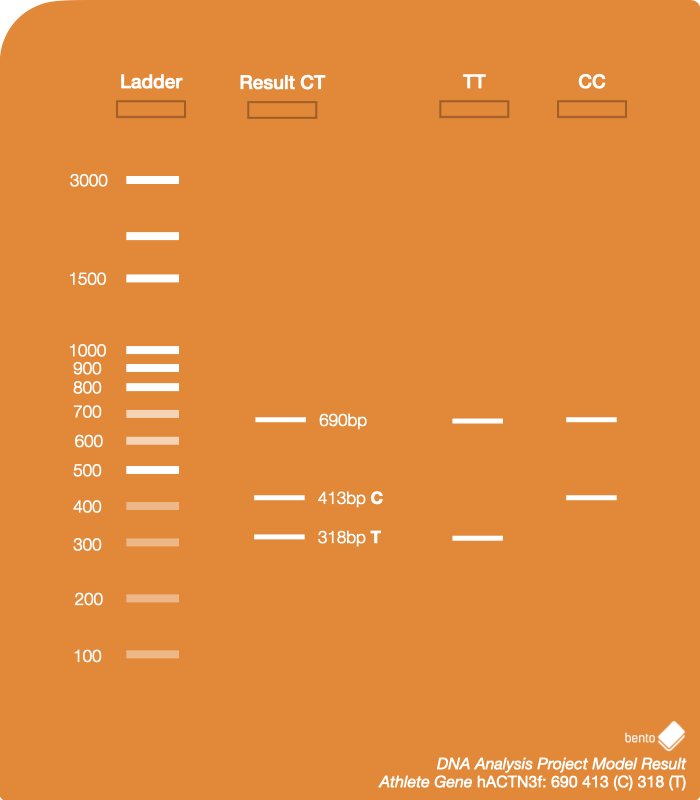
ACTN3 gene variations
You will explore two forms of the ACTN3 gene: The T allele and the C allele.
- C allele is associated with full power skeletal muscle contraction due to fast-twitching muscles fibres, which means the person could be an excellent sprinter.
- T allele is correlated with more efficient energy disposure due to slow-twitching muscles fibres, which means the person could have more endurance.
What are the possible results for this experiment?
There are three possible results, since the gene has two variations, and two occurrences of the gene exist – one from each chromosome:
- Homozygous dominant: Two copies of C allele, which means the person could have better performance as an sprinter.
- Homozygous recessive: Two copies of T allele, which means the person could have better endurance.
- Heterozygous: One copy of the C allele, and the T allele. The C allele is dominant over the T allele, so the effect on muscles performance will be similar to the homozygous dominant case.