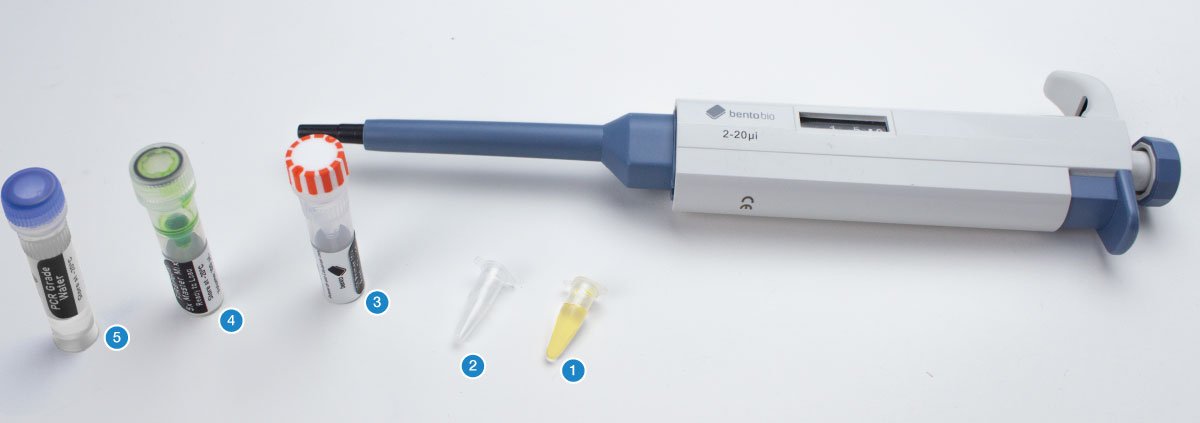
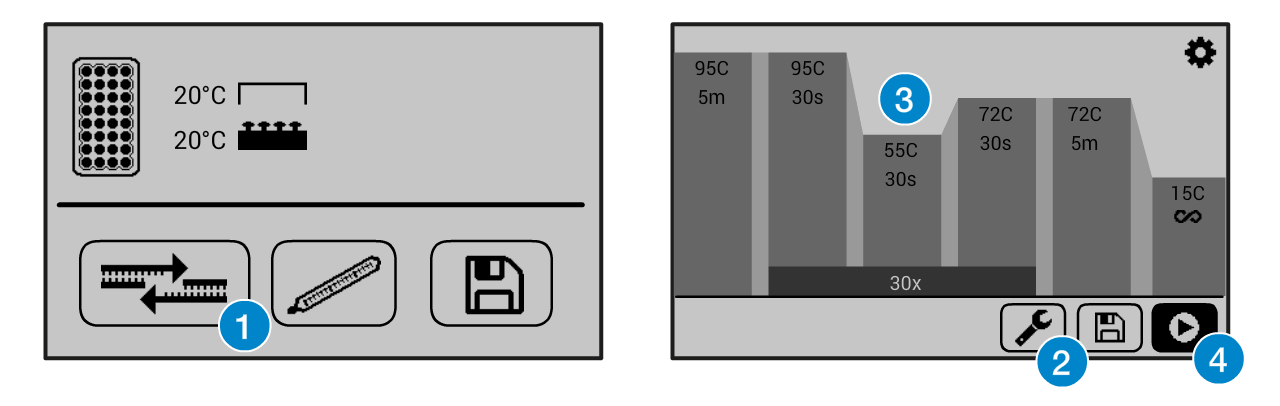
Overview
TAS2R38 gene variations
You can taste bitterness flavours thanks to your tongue receptors. One of these receptors is the Taste Receptor 2 Member 38, which is responsible for detecting PTC. The presence of this receptor is determined by the gene TAS2R38. You will explore two forms of the TAS2R38 gene: The T allele and the C allele.
C allele is associated with the presence of the receptor, which means you could taste PTC and bitterness.
T allele is associated with the absence of the receptor, which means you could not taste PTC and bitterness.
What are the possible results for this experiment?
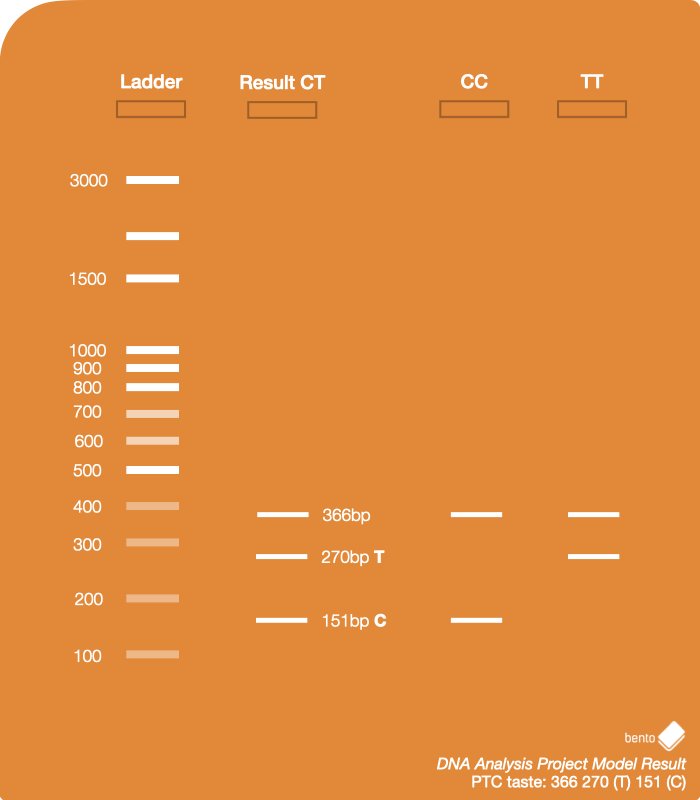
There are three possible results, since the gene has two variations, and two occurrences of the gene exist – one from each chromosome:
- Homozygous dominant: Both copies of the gene contain the C allele, which means the person can taste bitterness.
- Homozygous recessive: Both copies of the gene contain the T allele, which means the person is likely to be unable to taste bitterness.
- Heterozygous: One copy of the gene contain the T allele, and the other is C allele. The C allele is dominant, so the person likely to be able to taste bitterness.