Hi PCR enthusiasts!
Here are some recent highlights from Bento Lab.
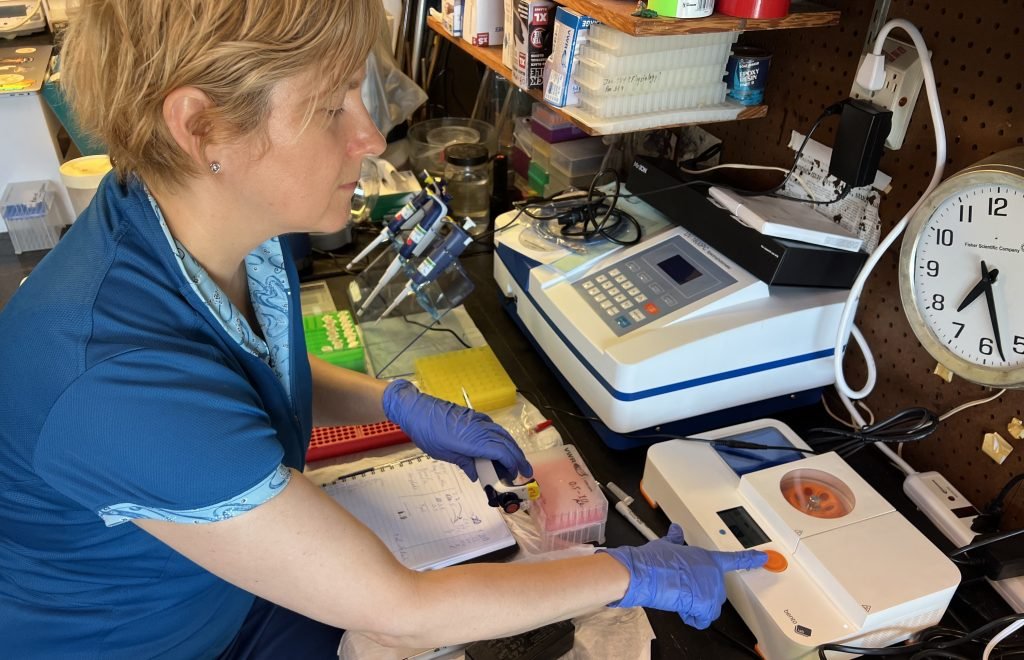
Bento Lab in the field
Unlocking the Mysteries of the Wild Ocean
We recently had the privilege of talking to Dr. Cheryl Ames, Professor of Applied Marine Biology, Tohoku University, Japan, about her exciting research.
Dr. Ames, an expert in jellyfish biodiversity and genomics, is making it possible to predict the presence of specific marine species in different regions of the ocean. Does today look like a cloudy day at the beach, with a high chance of deadly jellyfish?
Dr Ames wants to make such predictions a reality. She uses Bento Lab as a field PCR workstation and for quick and convenient troubleshooting in her lab.
Read the whole story about Dr. Ames’s awesome work here.
Interested in DNA methods and workflows? Subscribe for monthly insights.
Resources for Learners
We’ve been busy adding new articles to cover some of the obstacles faced by beginner (and even more experienced molecular biologists)
We would love to hear your feedback on these! Is anything good / bad, too long / too short, inspiring / boring, too simple / too complicated? Let us know how to support you better, and please feel free to send us your thoughts.
And if you have any particular requests for resources that would be useful to you or your colleagues, please get in touch.
Papers we love
This is a proof-of-concept article producing a detection workflow for a genus of plankton that causes Paralytic Shellfish Syndrome, one of the four recognised syndromes of shellfish poisoning in humans.
The authors, from the Centre for Environment Fisheries and Aquaculture Science (CEFAS), UK, came up with a number of novel and inventive approaches to doing this work in the field, including:
- Using a modified orbital power tool for cell lysis from sea water samples
- Using recombinase polymerase amplification instead of PCR to amplify DNA quickly without PCR inhibition
- Developing their own specific primers for genus Alexandrium
- Oxford Nanopore MinION metabarcoding
- Developing a portable power-efficient computer for nanopore read processing
- And much more!
Innovations at every step!
Bento Lab was used for incubations, PCR, and centrifugation in many places throughout their workflows.
You can read the entire paper here.
Methods and Techniques: PCR-RFLP
We were inspired to take a look at PCR-RFLP by a recently published five-year study of mammal biodiversity in a reserve in Moldova (Nistreanu et al. 2023, from a team associated with the Institutul de Zoologie, Universitatea de Stat din Moldova).
As a small part of this study, the researchers used Bento Lab to distinguish between wild boars from feral pigs using a PCR-RFLP assay (PCR followed by restriction enzyme digests and gel electrophoresis). Read the paper here (it’s mostly written in Romanian) and if you’re interested in the protocol you can check out the article where it was first published.
RFLP (restriction fragment length polymorphism) is a “DNA fingerprinting” technique whereby a DNA region is amplified and cut using enzymes at a specific nucleotide motif. The pattern of fragment sizes can be used to characterise and/or identify the sample when analysed by gel or capillary electrophoresis.
Wikipedia claims that RFLP is a “largely obsolete” method, but we were able to find a wide range of PCR-RFLP applications used as recently as 2023, such as in bird sexing; parasite detection; identifying different animals (such as ghost crabs in this article); rapid screening of fungal cultures; differentiating coffees of different origins, and authenticating grape varieties used in wine.
We think that PCR-RFLP could be a powerful tool for people wanting to do similar work to the above with Bento Lab, as a step up from conventional PCR assays without requiring the additional costs of DNA sequencing.
What do you think?
Looking for advice on using Bento Lab?
Book a free consultation or ask a question.We get many emails asking us: “When I buy a Bento Lab, what else will I need?”
In this post, we cover the main points to answer this frequently asked question.
The main factors to consider are:
- what you want to do with Bento Lab
- how much you want to stock up on reagents and consumables in your first purchase
Beginner’s Choice: Biotech 101 Kit
If you are a beginner interested in starting your DNA learning journey, one option is to buy Bento Lab together with the Biotech 101 Kit. This will have everything you need to run a small range of different learning experiments. It will give you a good introduction to the steps involved and give you a feel for where you can go next. See the list below for information on whether Bento Lab Entry or Pro is right for you.
If you want Bento Lab for specific applications, there is a core set of equipment and reagents that nearly everyone using Bento Lab will need. We have listed and described these below.
How much of each item should I buy?
In terms of volumes, we recommend buying enough of the consumable items for your planned use for the near future. We make reagents and consumables available at lower volumes so that you don’t need to break the bank on a year’s worth of supply. You can then stock up when you are running low on anything.
Working out the cost of getting started
A simple and quick way to calculate the overall budget you need is to simply add items to the shopping cart on our website as you read through. You can do this by clicking on the hyperlinked text for each product. You can also send the shopping cart to yourself as a PDF quote which you can order later. To do this, simply go to the cart and choose Request Quote instead of Purchase Now.
A Detailed Buyer’s Guide
From DNA extraction kits to plastic consumables, there are quite a few items to consider. We have grouped them into these categories so you can easily find what you are looking for on this list.
- Equipment
- Plastics Consumables
- Extraction Kits/Reagents
- PCR reagents and primers
- Gel electrophoresis reagents
Interested in DNA methods and workflows? Subscribe for monthly insights.
Equipment
1. Bento Lab
Entry or Pro – Which configuration is right for me?
- The Pro configuration is designed for ambitious and professional research applications. If you are using Bento Lab for real world applications or you need versatility, this is probably the right option for you.
The centrifuge is compatible with spin-column DNA extraction kits, and you can use it to precipitate and centrifuge DNA into pellets. The electrophoresis module has a variable voltage and can be used to run fast or very slow gels. - The Entry configuration is a more affordable option designed for beginners. It has a fixed voltage electrophoresis setup, and a slower fixed speed centrifuge.
If you don’t need a fast and variable centrifuge for faster or more ambitious DNA extractions, this option could be a great choice for you to get started.
If you are planning on taking your Bento Lab on fieldwork or to many different events on a regular basis, you may also want to invest in a carry case.

2. Adjustable micropipettes
You will need at least one micropipette to cover very small volumes for PCR. Another pipette to cover larger volumes for DNA extraction and other purposes is also highly recommended.
20 µL and 200 µL micropipettes do a good job of covering both ranges, and have the advantage of fitting the same pipette tips (2-200 µL) provided the tips are unfiltered.
You may also want to consider:
- A 1000 µL pipette (and tips) if you are routinely pipetting volumes larger than 200 µL
- A 10 µL pipette (and tips) if you want to have better control and accuracy for very small volumes
- A dedicated small pipette (20 µL or 10 µL) for handling PCR products only to reduce the risk of cross-contamination
- Other size micropipettes (2 µL, 50 µL, 100 µL, 300 µL) are available from other retailers if you find you need these volumes routinely
- A multichannel pipette if you are working with multiple Bento Labs or have particularly pipetting-intensive protocols

3. Add-A-Lane Gel Comb
You can double the number of PCR results you visualise in one gel by adding an additional row with an Add-A-Lane comb, and then running the DNA only half the way down the gel for each row.
This is very useful for when you want to visualise just a single band for each PCR reaction, but it can result in lower resolution for some applications compared to a full length gel run.

4. Additional equipment
You will also need some additional equipment, most of which you probably already have or can easily get:
- A microwave for melting agarose for gels
- A beaker (or a jam jar) for melting agarose
- A secure container for used pipette tips
- A secure container for implements used for sampling tissues, such as razor blades or needles — a sharps box would be ideal
- A plastic tray as a work surface — this can be bleached if needed
- Secure storage boxes to store equipment and reagents
- Plastic press-to-seal bags to store PCR products.
- Nitrile or latex gloves to protect your samples and PCR products from cross contamination
- Depending on your DNA extraction method you may need a method of grinding tissue — for example, to use the Dipstick DNA extraction kit we recommend plastic pestles for use with 1.5 mL centrifuge tubes
Plastic consumables
5. Pipette tips
You will need a LOT of pipette tips during your PCR journey! For example, an eight sample plus two controls single gel workflow using the HotSHOT DNA Extraction Kit uses a minimum of 27 x 2-200 µL pipette tips. After three such experiments you will have nearly used up a whole box of 96 tips.
When deciding what to buy, first make sure that your pipette tips fit the size ranges of your micropipettes:
- 20 µL and 200 µL pipettes can use 2-200 µL tips
- 1000 µL pipettes use 100-1000 µL tips
- If you choose to get 10 µL micropipettes, these use 10 µL tips
Secondly, you could think about how you can save plastic waste (and money). For example:
- Buy a few boxes of tips, and then get loose tips to refill the empty boxes. But note that some tip boxes and tips are not compatible. You may need some trial and error to find the right combinations.
- If you have a slow afternoon, you can look online for good deals on boxed tips. Sometimes you will find good deals for bulk buys from ex-lab stock.
You may also want to consider buying filtered pipette tips to help reduce the risk and impacts of PCR or reagent contamination, especially if your work is particularly valuable or sensitive. But be aware that filtered 20 µL and 200 µL tips are not interchangeable.
Reusing non-filtered pipette tips after washing and decontamination is technically possible, and is fine for practice pipetting where contamination doesn’t matter. But for PCR applications it is not usually economical for the average user, and it won’t be possible to reuse filter pipettes.
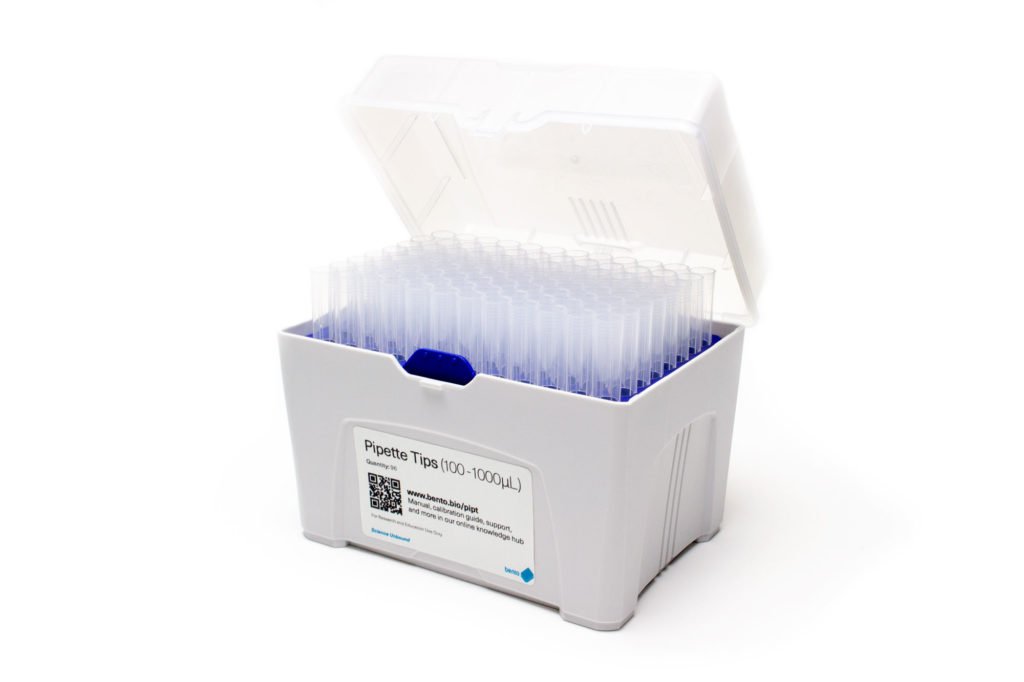
6. 1.5 mL centrifuge tubes
1.5 mL centrifuge tubes are needed for many experiments and especially for DNA extraction, for example with the Dipstick DNA Extraction Kit. They are not expensive and a large bag should last a long time.
If you intend to only use 0.2 uL PCR tubes for DNA extraction and PCR (for example using the HotSHOT DNA Extraction Kit) then you may not need any 1.5 mL tubes for this method.

7. 0.2 mL PCR tubes
0.2 mL PCR tubes are essential for PCR, but are also used for some small-volume DNA extraction protocols such as the HotSHOT DNA Extraction Kit.
They can come either as individual tubes, or as strips of 8 PCR tubes connected together (with or without individual lids), or as 96 well plates for larger laboratory thermocyclers.
If you are intending to routinely do a large number of samples with Bento Lab, it is worth considering sourcing 8 tube PCR strips with individual lids to help you keep your samples organised. These will usually be more expensive than individual tubes.
For higher throughput work, especially if involving dilution steps, you could consider 32 well PCR plate segments or cut-down 96 well PCR plates.

Extraction Kits and Reagents
Whatever the PCR application you will need some form of DNA extraction method. Some researchers may make up their own bespoke DNA extraction reagents, and others may prefer a DNA extraction kit of some sort.
8. DNA Extraction Kits
We have created two affordable DNA extraction kits to cover a wide range of applications:
The HotSHOT DNA Extraction Kit: a simple weak alkaline lysis and neutralisation method which is appropriate for many applications. This is suitable for quick crude extractions where you do not need to grind the specimen and where PCR inhibition by the sample is not a major problem.
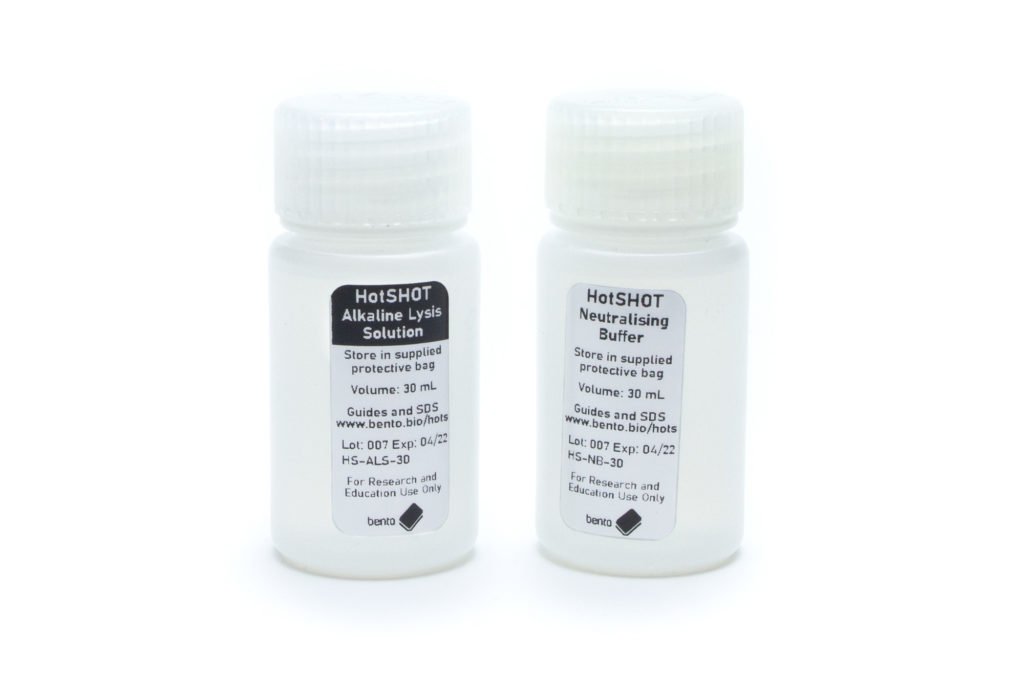
The Dipstick DNA Extraction Kit: a detergent/salt/buffer extraction method using filter paper dipsticks to transfer and wash DNA before transferring it into your PCR mix. This is also a quick method that allows you to grind the specimen to release more DNA while reducing potential PCR inhibition problems.

PCR reagents
To produce a PCR mix you will need three essential components:
9. PCR Mastermix
PCR Mastermixes are usually 5x or 2x working concentration. We offer two 5x PCR Mastermixes:
- 5x FIREPol® Master Mix Ready To Load — this is good for routine applications
- 5x HOT FIREPol® Blend Master Mix Ready to Load — this is a Hot Start polymerase, meaning that the polymerase enzyme is only activated after an initial heating step, thereby reducing the risk of primer dimer formation and non-specific amplification
For fieldwork applications you may prefer to buy multiple smaller volumes to reduce the risk of contamination or reagent loss when in the field.

10. PCR grade water
PCR grade water is highly purified water which is free from any contamination such as microorganisms, salts or solutes, or DNA and RNA-degrading enzymes. PCR grade water comprises a very large proportion of a PCR reaction so it is important for it to be clean.
Distilled water, or even good quality tap water, can be used for PCR and dilution steps, but at a risk of PCR contamination depending on any trace contaminants present and whether the primer set you are using will amplify these contaminants.

11. Primers
All PCR applications need primers (small strands of DNA matching the start and end of your amplification targets) to allow the PCR process to work. For every different PCR application you will need to buy specific primers for that application.
We supply primers as primer mixes for convenience, each containing the two or more primers needed for that PCR assay or barcoding application.
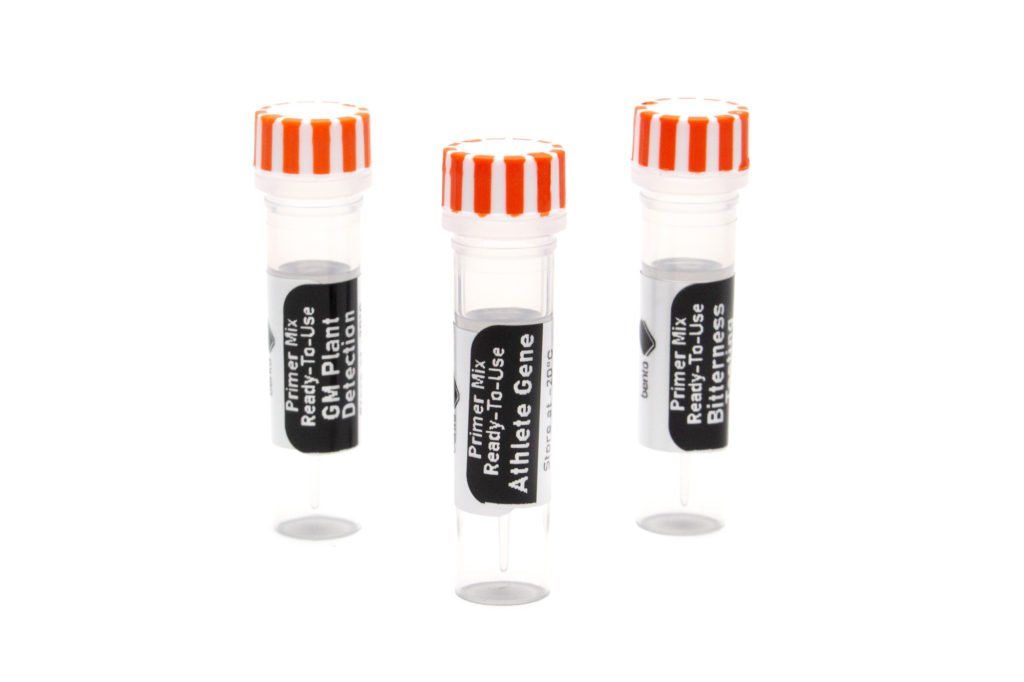
12. PCR additives
You may also want to add some PCR additives to help improve PCR efficiency or specificity, such as Bovine Serum Albumin (BSA), PCR-Enhancer solution, or Magnesium chloride. You should only buy these when you know you need them for your specific applications and how these reagents will improve you PCRs.
Gel Electrophoresis reagents
13. 10x TBE buffer
You will need 10x TBE Buffer to conduct electricity and to buffer DNA in electrophoresis gels and running buffer. We recommend using it at 0.5x concentration. 50 mL of 10x TBE will make and run 80 gels, and 1 L will make and run 400 gels.

14. Agarose Tablets
Agarose is needed to cast agarose gels for gel electrophoresis. We offer them as easy-to-use tablets. One tablet is usually used per gel, although you may wish to use two tablets for high-strength gels for better separation of 100-500 bp DNA fragments. You can buy tablets in strips of 10, or as a box of 200.
15. DNA stain
DNA stain is used to stain your PCR products or genomic DNA so that you can visualise them under blue light transillumination.
We recommend that beginners use GelGreen® DNA stain, which is a very bright and safe stain that can’t penetrate gloves or skin cells. It does have a tendency to smear when staining high concentrations of DNA stain due to the same mechanism that makes it safe – but this is a very sensible tradeoff to ensure safety.

16. DNA Ladder
DNA ladder is composed of fragments of DNA at fixed sizes so that it can be used as a reference for estimation of the size of your DNA strands. We recommend a 100 bp DNA ladder for most PCR assays, since most PCR products will end up within that size range.
We hope that this will give you an idea of what you will need to make the most of your Bento Lab!
Looking for advice on using Bento Lab?
Book a free consultation or ask a question.For those interested in the potential of PCR to combat foodborne illnesses and zoonotic diseases, Marin et al. (2022) present an innovative study where a portable sequencing kit — combining Bento Lab and the MinION nanopore sequencer (Oxford Nanopore Technologies) — was used for on-site detection and characterization of Campylobacter and other bacteria in approximately five hours.
“This method could provide a basis for future studies to help farmers implement active zoonotic bacterial screening in real-time diagnostics to take measures that will reduce the prevalence, which is indispensable for active surveillance.” – Marin et al. (2022)
The Challenge: Food Safety & Rapid Detection
Campylobacter is one of the most important foodborne pathogens globally, particularly in poultry. Early detection of Campylobacter outbreaks in broiler flocks could significantly reduce the risks of human infection. However, due to the rapid production cycle of broiler chickens (from egg to supermarket in ~43 days) and the scale of poultry farming, outbreaks need to be identified using fast, practical, and cost-effective methods.
Unfortunately, current methods of Campylobacter enumeration using microbiological culturing are time-consuming and can take up to five days of work. This delay from sampling to results is not ideal given the rapid production cycle and the fact that broiler chickens can be on supermarket shelves within 24 hrs of slaughter. Faster, sensitive, and cost-effective solutions are urgently needed.
The Study: A Proof-of-Concept Workflow
In this study, the authors (Marin et al., 2022) proposed and tested a proof-of-concept same-day, on-site Oxford Nanopore sequencing workflow to detect Campylobacter in broiler chickens, using Bento Lab as a portable PCR workstation and ONT MinION for sequencing.
The study involved 42 broiler chicks, with 20% being orally infected with Campylobacter. Caecal (gut) samples were taken at two key points:
- Day 14 – When the broiler immune system matures
- Day 42 – When the birds are taken for slaughter
Two detection workflows were compared:
- Standard microbiological culturing (ISO/TS 10272-2:2017)
- Portable Bento Lab & MinION sequencing workflow
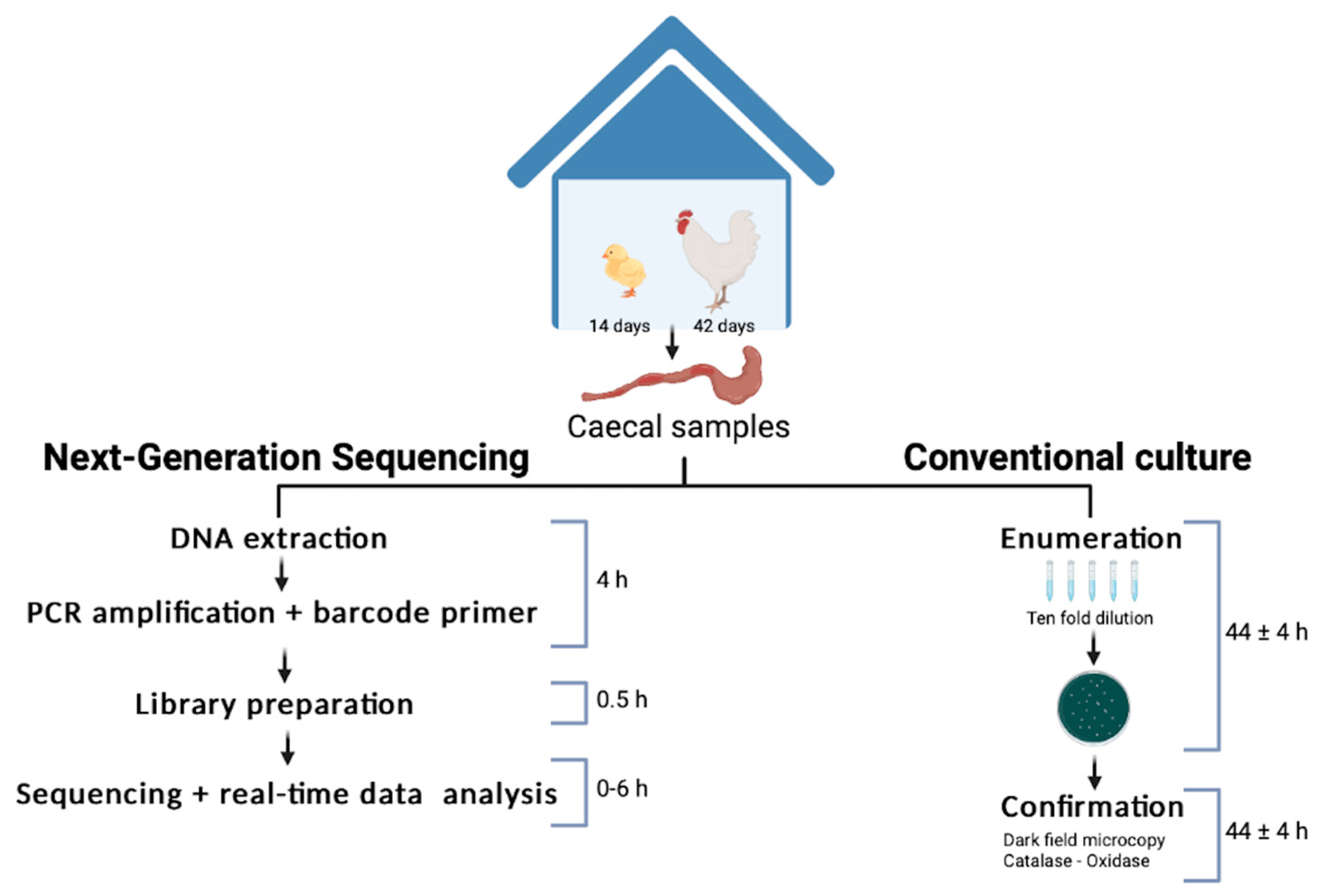
Timeline for the rapid method developed at farm level compared with the current culture detection method. Image CC-BY-4.0 the authors of Marin et al. (2022).
Bento Lab was used for
- DNA extraction from caecal contents using the Wizard Genomic DNA Purification Kit, Promega)
- Visualisation of DNA concentration and quality via agarose gel electrophoresis
- Library preparation for MinION sequencing
Following library preparation, the libraries were run on a MinION flow cell for six hours, with reads logged every 10 minutes to allow determination of the shortest amount of time needed to generate sufficient data for analysis. Raw reads were base-called using Albacore 2.1.7, poretools 0.06, and reads were assigned to taxonomic identities using RDP classifier.
The results showed that:
- Campylobacter was detected in all caecal samples, but it was absent in environmental samples and day-old chicks, demonstrating the sensitivity of the workflow
- The culturing ISO/TS 10272-2:2017 method took four days from start to finish, whereas the Bento Lab/MinION process produced actionable data within five hours.
- The Bento Lab/MinION workflow allowed simultaneous detection of Campylobacter as well as an assessment of bacterial taxonomic diversity and relative abundance present within the caecal samples, showing clear differences between the two sampling times.
The authors calculated that costs per sample for MinION sequencing were ~18x more expensive than the conventional culturing method. However, this could be reduced per sample by increasing the throughput, since ONT sequencing becomes significantly cheaper as sample numbers increase. The authors also didn’t consider additional costs of staff time.
Marin et al. (2022) concluded that “[we] have demonstrated that MinION-based workflow at the farm level is viable and highly accurate by comparing it with ISO/TS 10272-2:2017.”
Overall, this looks like an amazing application of portable PCR diagnostics for zoonotic disease detection, and one that could be applied in many more areas of the food production industry!
You can read the article here:
Marin et al. (2022). Rapid oxford nanopore technologies MinION sequencing workflow for Campylobacter jejuni identification in broilers on site—a proof-of-concept study. Animals, 12(16), 2065.
https://www.mdpi.com/2076-2615/12/16/2065
This article was originally published by Wayne Powell on the Bristol BioDesign Institute blog, shared with permission.
As part of City Nature Challenge 2022, a team of PhD candidates at the University of Bristol have led hands-on, family-friendly public engagement activities explaining how the cutting edge science of environmental DNA (eDNA) sequencing can be used to identify local wildlife. DNA sequencing has been taking place in universities and research labs for half a century, but the Bento Lab – a portable, hand-held DNA lab – is allowing citizen scientists to study the DNA of living things around them. The University of Bristol team used a series of escape room-like activities to bring to life the processes involved in using the Bento Lab.

The activities took place in Queen Square, Castle Park and Tyntesfield in Bristol, and were led by (l-r) Matt Tarnowski, Claire Noble, Hannah Langlands, Harry Thompson and Rosie Maddock.
An activity introducing the microscopic world of biology at our fingertips
DNA can be collected from inside an organism’s cells in a process that is a bit like popping a balloon. This was the first task to participants had to do – the balloon had been filled with the letters G, T, C, A to represent DNA – and they then extracted the ‘DNA’ using a pipette. As it is hard to study DNA from so few cells, a polymerase chain reaction (PCR) is used to amplify a particular DNA sequence within the cells. Attendees had to figure out how the PCR process works using coloured dominoes, and all succeeded in understanding that it involves repeatedly doubling the targeted DNA.
This amplified DNA can be selected using gel electrophoresis, which was demonstrated using a noisy rain-stick. The final step is sequencing the DNA, and participants enjoyed donning a cape to give them the ‘superpower’ of being able to read DNA. Since these lab tasks would take hours with the Bento Lab, environmental samples that were brought along were taken away for identification, and attendees will be notified of their species once analysed.

Interactive activities brought eDNA sampling, PCR, the Bento Lab & DNA sequencing to life (l-r)
Putting the mobile eDNA lab to the test
Some creatures, such as fungi or microorganisms, can be hard to spot, yet play essential roles in the environment. The next stop for the Bento Lab is to assess fungal diversity at a site of ecological restoration. The Refungium at Coed Talylan in Wales aims to foster the highest diversity of fungi in the UK. The Refungium will be a refuge for native mushrooms, and studying the fungi there with the Bento Lab will inform restoration of this 15-hectare semi-natural woodland.
Would you like to host the eDNA lab?
We welcome opportunities to share this portable workshop, be it at a school, college, university or festival. With activities for all ages, this citizen science experiment has the potential to inspire the next generation of bioscientists and stewards of the environment. As well as learning how eDNA studies work, it opens up conversations about genetics, synthetic biology, ecology, microbiology, sustainability, climate change and responsible research. These themes mesh with the sustainability and climate change strategy recently shared by the UK’s Department for Education. If you would like to host the eDNA lab, please write to us.
Matthew Tarnowski, Claire Noble, Hannah Langlands, Rosie Maddock and Harry Thompson developed this project, which was funded by the EPSRC/BBSRC Centre for Doctoral Training in Synthetic Biology (SynBio CDT) Outreach Award.
One of the common questions we get asked is: “Is it difficult to take Bento Lab on a plane?”
The answer is: It’s not difficult at all! Before Covid postponed field trips and moved conferences online, we regularly took Bento Lab on the road. Hopefully travel for field research can make a return next year. To help you prepare, here are my top 5 tips for flying with your Bento Lab.
1. Treat your lab as a standard electronic device

I carry Bento Lab in my hand luggage, and treat it just like a laptop. On all of my flights, I have never experienced issues taking it onboard. Before going through the security checkpoint, place Bento Lab in a separate bin to go through the scanner.
You may even get a few curious looks. Bento Lab can definitely be a great conversation starter! We have met security staff with a forensics background, and fellow molecular biologists on their way to conferences keen to compare notes.
2. Store portable batteries in hand luggage
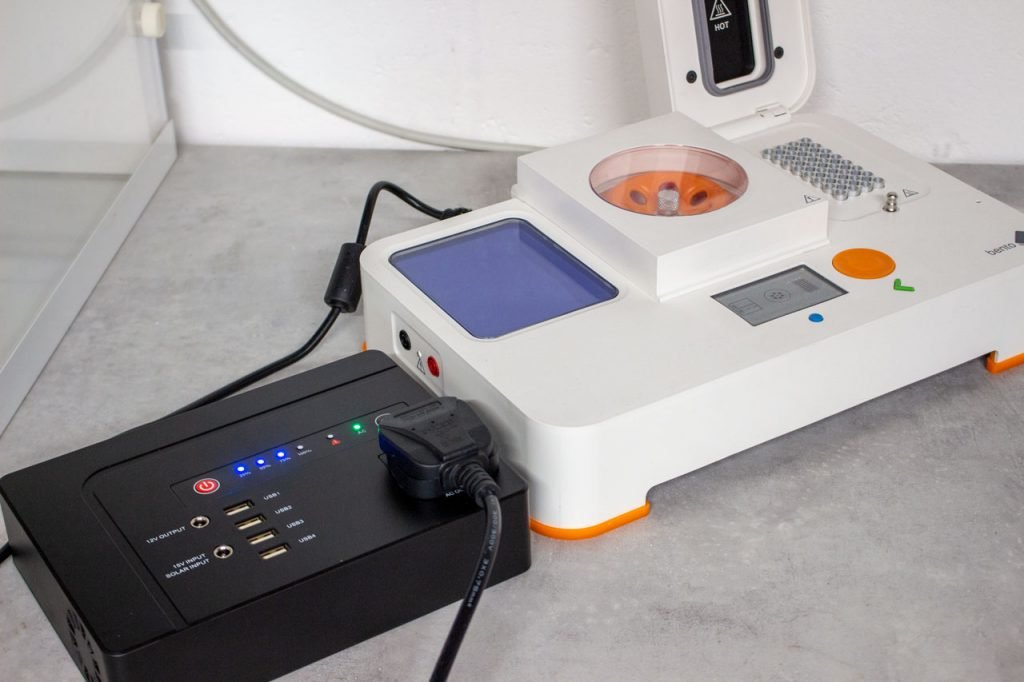
If you are travelling somewhere remote, you can use Bento Lab with portable batteries (also called power banks).
You will need to take the battery in your hand luggage. There are limits on what batteries you are allowed take on planes. Capacities of up to 100Wh are the most straightforward, and you are allowed to increase the capacity up to 160Wh if you notify the airline and get approval. Make sure that the battery is labelled with the capacity (e.g. 160Wh).
3. Protect your lab with a carry case

Bento Lab has a robust design, and I often carry mine in my backpack. But if you want to be safe, or you are going to very remote places, you can protect Bento Lab with a hard carry case. You can find a variety of foam-filled carry cases, and adapt the inside to suit your needs.
You can find a 20 L case for Bento Lab in our online store, and we even have a 34 L option with extra space for pipettes and tubes.

4. Travel size reagents
There’s no need to lug litres of buffer in your luggage, and some reagents don’t need to be frozen. I travel with a make-up size bag of reagents. My favourite travel friendly reagents are:
- Agarose tablets: These tablets come in 0.5g format. No need for scales, and fiddly power. You can make a gel with 1 tablet and 25mL of buffer.
- HOT FIREol® Blend Master Mix: This mastermix can be stored at room temperature for up to 30 days, which makes it ideal for fieldwork.
- Concentrated TBE buffer: Fitting into your liquid allowance, a 50 mL bottle of buffer at 20X concentration makes 1L of 0.5X working solution.
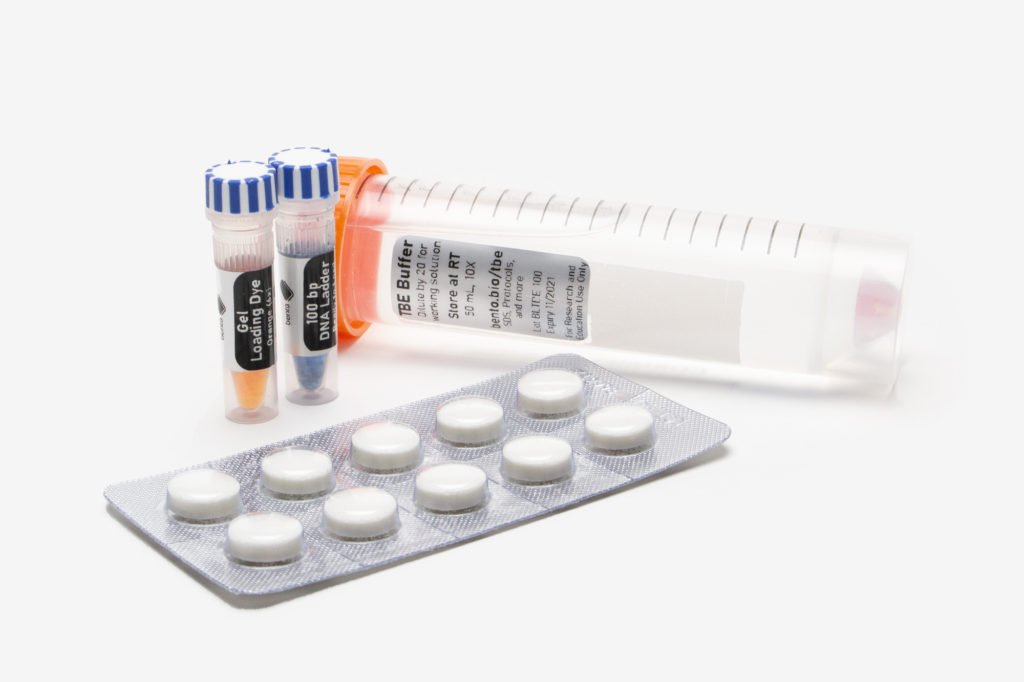
A great tip I picked up from my colleague Brian: Avoid contamination while you’re in the field by using aliquots. Divide mastermix into separate tubes, or buy several small 100 µL tubes instead of a bigger 1000 µL format.
If you are on a trip where taking Bento Lab is not possible, you could still use the Dipstick equipment-free DNA sampling method. DNA captured with dipsticks can be stored in TE buffer until you are home.
5. Download a letter for customs
For those concerned about questions at customs or security, we have a downloadable letter stating the purpose and product code of Bento Lab. You can download the letter here in the resource centre.

Bonus tip: Send us a virtual postcard!
We love seeing pictures of Bento Lab being used all around the world. Email us, or tag us on Twitter or Instagram!

Identifying the sex of birds can be essential for bird owners, breeders, conservationists, and many other people. But for many birds, it can be difficult or impossible to differentiate their sex based on appearance. A DNA test to determine the bird’s sex is often the best solution
Many avian sexing services exist which will analyse the DNA based on a drop of blood that can be sent in the mail. This is convenient, but can be take time to turn around, and gets expensive for large number of samples.
We’re excited to launch a new set of bird sexing resources for Bento Lab, that allow anyone to conduct their own DNA-based bird sexing.
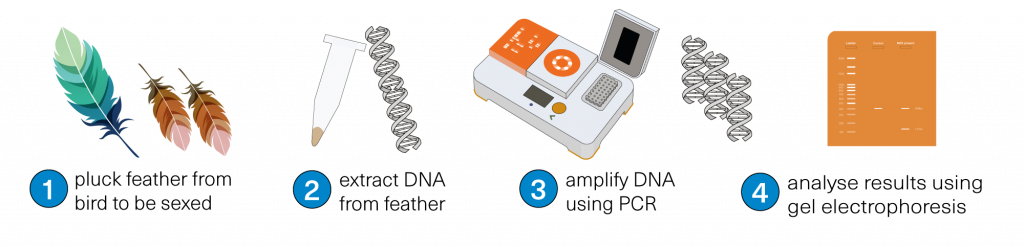
Our bird sexing workflow uses the HotSHOT DNA Extraction kit to extract DNA from feathers. Once the DNA is extracted, we have two sets of primer mixes to conduct PCR and gel electrophoresis.
You can find out more on the bird sexing page here.
Interested in getting started? Tell us what you need
We are passionate about unlocking the power of DNA analysis and making it accessible to anybody.
If you are thinking about trying bird sexing yourself, or if you are already conducting your own bird sex DNA tests, or using external bird sexing services – we would love to hear from you.
Everything you will need for this DIY bird sexing workflow is readily available. If you already have a setup for PCR and gel electrophoresis (such as Bento Lab), you can find both sets of bird sexing primers in our shop.
News from the Bento Team
The bird sexing resources were a project started by Brian Douglas, who was our first full-time molecular biologist.
Brian joined us on the heels of the Lost and Found Fungi project. He was instrumental in launching our supplies shop, and pioneering our DNA extraction kits.
He has now started an exciting new role at Kew Gardens, and although we’ll miss him, we’re also excited for him! We know his new project will make a splash. Thank you for your awesome work Brian!
The bird sexing resources started with Brian, but they were finished by our newest team member, Jenny.
She joined our team this month as our new molecular biologist! Watch this space – we’ve got a lot of exciting things planned.
Small but impactful hardware updates
We are always improving Bento Lab to make it more versatile and robust. Bento Lab now ships with a foldable Gel Imaging Hood, and Bento Lab Pro comes with an updated rotor design for higher capacity.
Gel Imaging Hood
The new Gel Imaging Hood is now included with Bento Lab, and will make it much easier to see and photograph gel bands in bright environments. The simple dark box design can fold flat, so that it takes up little space when you’re travelling. It includes an amber filter, so you can capture sharp pictures of your gels quickly and consistently.
2 mL Bento Lab Pro Rotor
We have updated the design of the centrifuge rotor for Bento Lab Pro, and it is now compatible with 2 mL tubes. The previous version of the rotor was limited to a maximum tube size of 1.5 mL unskirted tubes. This meant that commercial extraction kits such as Qiagen spin columns had to be modified to fit. With the new 2 mL rotor, these types of spin columns fit without modifications. You can read more about compatible tube sizes in our manual.
We’re excited to announce the release of a new software version for Bento Lab. The update improves stability, and brings new features that have been requested by user feedback.
You can see watch a walk through of the software changes on our YouTube channel here. If you are not yet a subscriber, our YouTube channel is a great way to stay connected.
We highly recommend all Bento Lab users to update their firmware following these steps. Firmware update can be launched automatically, when Bento Lab is connected to Wifi.
Run multiple modules
We have completely rewritten the power management software to enable running the PCR module concurrently with either the centrifuge or gel electrophoresis module. With the increasing popularity of LAMP workflows, we believe that this will be particularly useful for users who want to run the heat block at a fixed temperature whilst using the centrifuge for quick or sustained spins.
Additional changes and fixes
- Fixing of graphical glitches
- Improved early detection of heat block hardware issues
- 3 second waiting period until gel power is paused on disconnection
- Lid temperature can be customised
Installation
We highly recommend all Bento Lab users to update their firmware following these steps.
If you encounter any issues during your firmware update, please get in touch with us.
Download the official press release here.
We are very excited to announce that Bento Lab has won an award at this year’s Good Design Award.
The annual Good Design Awards is Australia’s oldest and most prestigious international Awards for design and innovation with a proud history dating back to 1958.
“This is a really fantastic and innovative product. Bento Lab democratises the world of genetics testing as it makes available research tools to emerging nations and educational users. Overall and inspirational design providing accessibility and education for many humans. Congratulations.”
The Good Design Awards Jury

Bento Lab received a Good Design Award Gold Accolade in the Product Design Medical and Scientific category.
More than 55 Good Design Awards judges evaluated each entry according to a strict set of design criteria which covers ‘good design’, ‘design innovation’ and ‘design impact’. Projects recognised with a Good Design Award must demonstrate excellence in good design and convince the Jury they are worthy of recognition at this level.
“Receiving a Good Design Award is a significant achievement given the very high calibre and record number of entries received in 2020.”
Dr. Brandon Gien, CEO of Good Design Australia
The Good Design Awards Jury praised Bento Lab, commenting: “The aesthetics of the technology are really playful and this will help to increase the accessibility of these sciences to those who may have been formerly discouraged from participating. The product can have a great deal of impact in parts of the world where genetic testing is not readily available. Reducing cost, providing educational and support materials and ensuring accessibility are steps forward to increase adoption of genomics around the world. The designers have addressed this need in a “field kit” capable of being cheaply deployed in remote areas or the field, moving the product closer to the “point of interaction”.
We are honoured to have received this award, and the whole Bento Lab team is proud that our mission of creating accessible tools for biosciences has been recognised by the Good Design Award jury.
Thank you to the jury, and to everyone who has supported us on our way!

Watch our interview with Athanasia Dourou on our YouTube channel.
When it comes to paying a premium for extra virgin olive oil, how do you know that you are getting what’s on the label? This week we spoke with Dr Athanasia Dourou, the head of research and development at BioCoS, a Greek start-up.
Based on the island of Crete, the company is using DNA to tackle food fraud. They have validated their technology, focusing initially on testing the authenticity of olive oil.

Photo: BioCoS
In 2016, Forbes reported that as much as 80% of olive oil on the market is fraudulent or mislabelled. Along with Italy and Spain, Greece is the third largest producer of olive oil in the world. Premium oils, including the favourite extra virgin olive oil, are particularly at risk of adulteration by cheaper vegetable oils for illegitimate profit.
Traditionally olive oils are identified based on chemistry, using a type of spectroscopy called Nuclear Magnetic Resonance (NMR). However, determining the geographical origin of olive oil relies on many different components. The chemical composition of an olive oil is a result of different factors such as climate, genotype, and agricultural practices. Among researchers, how to define the exact origin is controversial.
Using a tailor-made bioinformatics tool, BioCos has identified novel biomarkers to determine olive varieties, validated at the University of Wisconsin-Madison, in the USA and the IBBR-CNR in Perugia, Italy. The biomarkers allow the team to create a DNA fingerprint that is unique to the type of olive, enabling them to distinguish between 19 different varieties.
We are able to identify the different varieties with 95 – 97% confidence.
Athanasia-Maria Dourou
The team works directly with premium olive oil producers that source oil from different producers. To get a competitive edge, these companies are looking to demonstrate to consumers that their products are authentic.
To validate the biomarkers they have developed, Athanasia extracts DNA from leaf samples or olive oil, and compares the samples to other plant species, such as grape seed or quinoa relatives. Then, she compares different varieties within the same species using sequencing, generating a final DNA fingerprint to identify the olive oil and demonstrate authenticity.
Bento Lab is very convenient, especially as a startup. Having equipment that is both compact, and combines all the devices you need is important. And I like the setup!
Athanasia-Maria Dourou
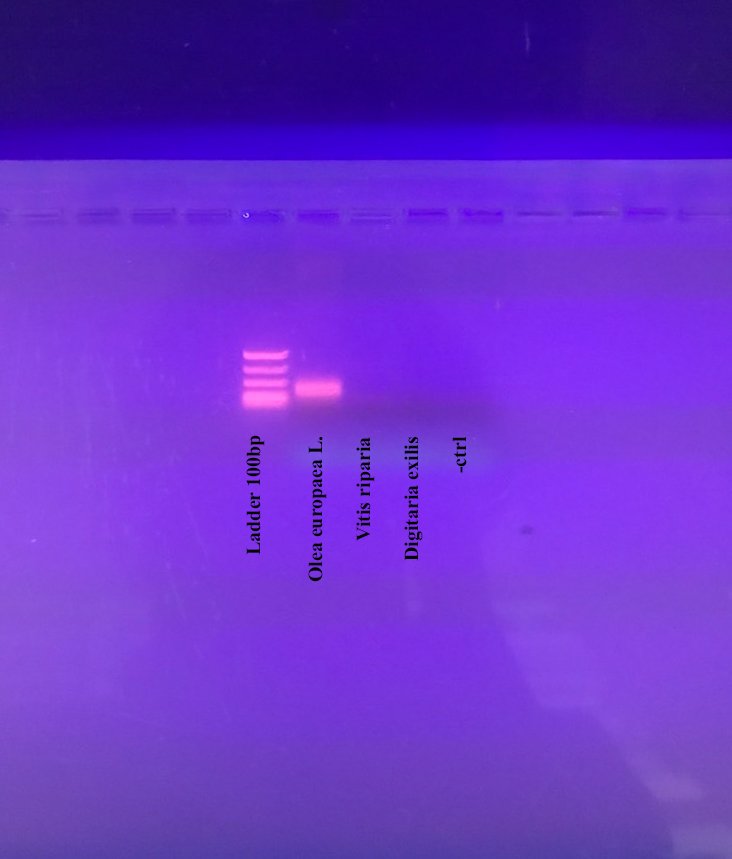
Moving from academia to start-up, Athanasia says the biggest challenge was switching into a business mentality. Their clients are interested in the results, rather than the scientific process. In industry, more attention is paid to managing the clients’ needs and expectations – a critical component of managing relationships, which is given little attention in academia.
BioCoS were recently awarded funding through the Horizon 2020 programme for SME Instrument Phase 1, a fiercely competitive EU grant. In September they will begin applying their technology to health applications, including SARS-CoV-2, the Ebola virus and foodborne diseases.
As a result of their success, BioCoS are growing. They are currently recruiting for 4 positions, covering bioinformatics, molecular biology, marketing, and project management. Their founder, Dr Stelios Arhondakis, advocates strongly for building the local innovation economy in Crete, says Athanasia. He is keen to demonstrate that young scientists can find meaningful technology jobs and innovate at home in Greece.
Find out more about Biocos and their openings on their website. They share regular updates on LinkedIn, and you can also keep up with the team by following on Twitter and Facebook.
Come work with us in @BioCoS_! We are an emerging Biotech company located in Greece, and as our company grows, we are looking for 4 highly motivated people to join our team!
— BioCoS (@BioCoS_) July 13, 2020
For more information click on the link belowhttps://t.co/6ibfcqa4uz pic.twitter.com/ndH0P19CJm



